Translate this page into:
Detection of transmembrane protein 100 in breast cancer: Correlation with malignant progression and chemosensitivity

*Corresponding author: Ling Zhao, Department of Ultrasound, Zibo Central Hospital, Zibo, Shandong, China. 18560291381@163.com
-
Received: ,
Accepted: ,
How to cite this article: Liu X, Zhang G, Zhao L. Detection of transmembrane protein 100 in breast cancer: Correlation with malignant progression and chemosensitivity. CytoJournal. 2024;21:65. doi: 10.25259/Cytojournal_107_2024
Abstract
Objective:
With increased incidence, breast cancer has become the most common malignant tumor in women. Transmembrane protein 100 (TMEM100) is a key factor affecting the progression of malignant tumors. The aim of the study is to examine the molecular mechanism of TMEM100 in malignant progression.
Material and Methods:
TMEM100 expression was analyzed by Western blot, immunohistochemistry, and real-time quantitative polymerase chain reaction. Cell migration and invasiveness after transfection with TMEM100 were investigated by Transwell assay. 5-ethynyl-2-deoxyuridine staining and cell colony-formation assay were utilized to the exploration of cell proliferation. Flow cytometry was adopted to detect whether TMEM100 affected the effect of Docetaxel on cell apoptosis. The effects of TMEM100 on the Ras-extracellular signal-regulated kinase (RAS/ERK) pathway were explored by Western blot assay.
Results:
Downregulated TMEM100 expression was in breast cancer tissues (P < 0.01). TMEM100 overexpression hindered the invasion (P < 0.01), migration (P < 0.01), and proliferation (P < 0.01) of breast cancer cells. Chemotherapy sensitivity of breast cancer cells to docetaxel was enhanced by TMEM100 (P < 0.01). TMEM100 inhibited Ras expression and ERK1/2 phosphorylation (P < 0.01). Furthermore, ERK agonist TertButylhydroquinone neutralized the effects of TMEM100 (P < 0.01).
Conclusion:
TMEM100 blocked malignant progression of breast cancer and enhanced docetaxel chemosensitivity by suppressing RAS/ERK pathway. These data manifested that regulation of TMEM100 expression may affect the progression of breast cancer, and its prognostic value and mechanism deserve further investigation.
Keywords
Transmembrane protein 100
Breast cancer
Chemosensitivity
Ras-extracellular signal-regulated kinase pathway
INTRODUCTION
Breast cancer has currently overtaken lung cancer to become “the number one cancer in the world.”[1] Breast cancer is more common in middle-aged female friends over the age of 40.[2] Chemotherapy, endocrine therapy, and targeted therapy are the most important drug treatments for breast cancer at present, effectively reducing the risk of death and recurrence.[3] However, some patients may lack effective therapeutic targets, have poor therapeutic effect, or develop resistance after repeated drug treatment, resulting in poor prognosis.[4] The most widely used chemotherapy of breast cancer is taxane chemotherapeutic drugs.[5] Docetaxel, as a chemotherapeutic drug of purpurane, can cause patients to have intrinsic drug resistance or gradually develop acquired drug resistance during treatment, leading to cancer recurrence and metastasis.[6] Therefore, identifying new biomarkers and therapeutic targets is necessary for predicting the prognosis of breast cancer patients and developing new clinical-treatment strategies.
As a new family of membrane proteins, the transmembrane protein (TMEM) family is extensively involved in basic biological functions, including material transport, signal transduction, and as a basic component of the membrane. For human cancers, TMEM family proteins play tumor-suppressor and oncogene roles. They are related to tumor progression and invasion, as well as chemotherapy resistance.[7] Multiple TMEM proteins have been observed to suppress and promote the development of breast cancer. For instance, TMEM17 exacerbates the malignancy of breast cancer cells.[8] TMEM178 downregulation is associated with promoter hypermethylation, and evidence shows a correlation with breast cancer patient prognosis, pazopanib sensitivity, and tumor immunity.[9] TMEM100, originally identified by Kawai et al. in the mouse genome, comprises 134 amino acids and is highly conserved in vertebrates.[10] Functional studies of TMEM100 have focused on its role in blood-vessel formation.[11,12] TMEM100 participates in the regulation of human lymphatic endothelial cells in the main vein.[13] TMEM100 plays a pivotal role in nervous system and pain, transmitting pain signals.[14] As an anti-oncogene, the mutation or abnormal expression of TMEM100 may lead to genomic instability, thereby promoting tumor formation.[15] The expression level of TMEM100 is relatively low in esophageal cancer tissues, and its overexpression hinders the growth, and metastasis of esophageal cancer cells.[16]
The aim of this study was to investigate the function of TMEM100 on the malignant progression of breast cancer cells in vitro. The mechanism of TMEM100’s involvement in breast cancer through the RAS-extracellular signal-regulated kinase (RAS/ERK) pathway was studied. At the same time, the role of TMEM10 on the sensitivity of docetaxel was also investigated.
MATERIAL AND METHODS
Clinical data
Cancerous and paracancer tissues of 40 breast cancer patients treated in Zibo Central Hospital were collected. The tissue samples were surgically removed, stored in liquid nitrogen, frozen overnight, and transferred into a cryogenic refrigerator at −80°C for storage. All experiments were performed in accordance with the ethical guidelines of Helsinki Declaration and with the informed consent of all patients. This research was funded by the ethics committee of Zibo Central Hospital (No. 2024129).
Cell clines
All cells were purchased from Pricella (Wuhan, China) and identified through mycoplasma detection and short-tandem repeat. Dulbecco’s modified Eagle’s medium (CL-B874, YiHe Biotechnology, Shanghai, China) was used to culture MCF-7 (CL-0149), MDA-MB-231 (CL-0150), and MCF-10A (CL-0525). Roswell Park Memorial Institute-1640 medium (PM150110, Pricella, Wuhan, China) was used to culture BT474 (CL-0040), T47D (CL-0228), and SK-BR-3 (CL-0211). The cell culture conditions were set to 37°C and 5% carbon dioxide.
The TMEM100 overexpressed plasmid (pcDNA-3.1 + TMEM100) and the corresponding empty vector control plasmid (pcDNA-3.1) were designed and provided by Heyuan Bio Corporation (Shanghai, China). The coding sequences of TMEM100: ATGACTGA AGAGCCCATCAAGGAGATCCTGGGAGCCCCAA AGGCTCACATGGCAGCGACGATGGAGAAG AGCCCCAAGAGTGAAGTTGTGATCACCACAGT C C C TC TG G TC AG TG AG AT TC AG T TG AT GGCTGCTACAGGGGG TACCGAGCTCTCCTGCTACCG CTGCATCATCCCCTTTGCTGTGGTTGTCT TCATCGCCGGCATCGTGGTCACCGCGGTGGCTT ACAGCTTCAATTCCCATGGGTCTATTATCTCCATCT TTGGCCTGGTTGTTCTGTCATCTGGACTTTT TTTACTAGCCTCCAGTGCCTTGTGCTGGAAAGT G A G A C A A A G G A G C A A G A A A G C C A A GAGACGGGAGAGTCAAACAGCTCTCGTGGCA AATCAGAGAAGCTTGTTTGCTTGA. The transfection was performed using Lipo3000 (L3000015, Invitrogen, United States of America) transfection reagent when the cells grew to 70%. About 48 h after transfection, TMEM100 expression was detected by Western blot assay and real-time quantity polymerase chain reaction (RT-qPCR).
Cytotoxicity of docetaxel
We tested the effect of docetaxel (114977-28-5, MCE, New Jersey, United States of America) on cytotoxicity through the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. We inoculated 100 ìL of cells (5 × 104 cells/mL) onto 96-well plates and divided them into a control group and docetaxel groups (2, 4, 8, 16, 32, and 64 nM). MTT method (ml094804, enzyme-linked, Shanghai, China) was used to measure the absorbance at 570 nm on a microplate reader (YP-96B, YouYunPu, Weifang, China).
Cell colony-formation assay
Single suspension cells were prepared by digestion of 0.25% trypsin (S10032, Yesen, Shanghai, China). 2 mL cell suspension was added onto the six-well plate for routine culture. The culture was terminated when clone white spots were visible in the petri dish. 2 mL of 5% paraformaldehyde (P0099, Solarbio, Beijing, China) was added for 15 min. Then, cells were stained for 10–30 min with1 mL of 0.5% crystal violet solution (548-62-9, Abiowell, Changsha, China). Finally, the number of cell clones was counted.
5-Ethynyl-2-deoxyuridine (EdU) staining
2 × 104 cells per well were inoculated onto a 96-well plate and cultured with 100 μL of EdU solution (50 μM; CA1170, Solarbio, Beijing, China) for 4 h. Each well was fixed by adding 50 μL of PFA (PN4205, G-CLONE, Beijing, China) for 30 min and decolorized with 50 μL of glycine (G8200, Solarbio, Beijing, China) for 5 min. Then, 100 μL of penetrant (0.5% TritonX-100, T9284, Sigma Aldrich, United States of America) was added for 10 min. For staining, 100 μL of Apollo and 100 μL of ×1 Hoechst33342 (CA1170, Solarbio, Beijing, China) were added. The observation of cells was using a microscope (DSY5000X, Chongguang, Chongqing, China).
Flow cytometry
After digestion, the cells were centrifuged with 1 mL PBS (BC-PBS-01, Biochannel, Nanjing, China) at 4°C. After fixation for 24 h, cells were stained with Annexin V-Fluorescein isothiocyanate/Propidium iodide kit (40302ES60, Yesen, Shanghai, China). The apoptosis rate of different treatment groups was measured with a flow cytometer (CytoFLEX, Beckman Coulter, United States of America).
Transwell assay
For the Transwell invasion experiment, 50 μL of Matrigel (M8370, Solarbio, Beijing, China) was placed on a 24-well plate containing 500 μL of medium. The upper chamber was inoculated with 3 × 105 cells of logarithmic growth stage. After fixation (4% paraformaldehyde: PH0427, Condice, Wuhan, China), cells were stained (0.5% crystal violet: C0121, Beyotime, Shanghai, China) for 15 min. The cells exiting the chamber were counted by a microscope (DM1600HD, Leica, Germany). The Transwell migration experiment was similar to the invasion experiment, but the chamber did not need to be coated with Matrigel.
Immunohistochemical (IHC) staining
Paraffin sections were dewaxed for hydration and antigen repair. Paraffin sections were incubated with 10% goat serum sealing solution. Then, tissue sections were incubated with primary antibody (1:100, abs112915, Absin, Shanghai, China) and second antibody (ab6721, 1:1000, Abcam, Cambridge, United Kingdom). Diaminobenzidine (LM-0323, LMAI Bio, Shanghai, China) chromogenic reagent and hematoxylin (H3136, Sigma–Aldrich, United States of America) were added to stain for 30 s. After dehydration with ethanol, the tissue sections were made transparent with environmental protection transparent agent for 20 min and then sealed with neutral gum. The distribution of TMEM100 in tissues was observed under a microscope (LSM900, Zeiss, Oberkochen, Germany).
Western blot assay
Total protein was obtained after the cells were lysed and centrifuged. The detection of protein concentration was according to the bicinchoninic acidprotein concentration kit (20201ES, Yeasen, Shanghai, China). About 50 μg of protein sample was subjected to gel electrophoresis and then transferred from gel onto polyvinylidene fluoride membrane (IPVH00010, Millipore, Boston, Massachusetts, United States of America), which was then placed in a sealing solution containing 5% skim milk powder. After adding primary antibody, the membrane was incubated overnight at 4 °C. After incubation in diluted secondary antibody, an ECL chemiluminescence kit (3622ES76, Yeasen, Shanghai, China) was used for luminescence detection on the chemiluminescence gel-imaging system (JY-Clear ECL, JunYi, Beijing, China). The gray value of the strip was measured using Image J (National Institutes of Health, New York, United States of America). The primary antibodies were anti-β-actin (1:10000, AC038, ABclonal, China), anti-TMEM100 (1:1000, A16653, ABclonal, China), anti-ERK1/2 (1:1000, A4782, GeneTex, United States of America), anti-RAS (1:100, GTX132480, GeneTex, United States of America), anti-p-ERK1/2 (1:1000, ab32538, Abcam, Cambridge, United Kingdom), anti-MEK (1:1000, ab32091, Abcam, Cambridge, United Kingdom), and anti-p-MEK (1:1000, ab96379, Abcam, Cambridge, United Kingdom). The secondary antibody was obtained from Abcam (1:5000, ab6721, Abcam, Cambridge, United Kingdom).
RT-qPCR
The extraction of total RNA was obtained through Trizol (15596-018, Solarbio, Beijing, China) According to reverse transcription kit (BTN63826, Baiao Laibo, Beijing, China), complementary deoxyribonucleic acid was obtained. After the reverse transcription was completed, the gene expression was analyzed by Applied Biosystems 7500 real-time polymerase chain reaction (Thermo Fisher Scientific, Waltham, Massachusetts, United States of America). Using β-actin as the internal reference gene., the relative mRNA expression of each gene was obtained according to 2−△△CT. The following table reveals the sequence of primers [Table 1].
| Primer name | Sequence (5′-3′) |
|---|---|
| TMEM100-Forward | CATGGCAGCGACGATGGAGAAG |
| TMEM100-Reverse | CCGGCGATGAAGACAACCACAG |
| β-actin-Forward | AGAGCTACGAGCTGCCTGAC |
| β-actin-Reverse | AGCACTGTGTTGGCGTACAG |
RT-qPCR: Real-time quantitative polymerase chain reaction, A: Adenine,C: Cytosine, G: Guanine, T: Thymine. TMEM100: Transmembrane protein 100
Statistical analysis
The experimental results were statistically analyzed using GraphPad Prism 8 (GraphPad Software Inc., La Jolla, California, United States of America). The P-value with statistical significance is <0.05. The comparison of multiple groups was performed by Analysis of variance, and the comparison of two groups was performed by t-test.
RESULTS
Downregulation of TMEM100 in human breast cancer tissues
The TIMER2.0 database shows the TMEM100 expression level in various cancer tissues. Results showed that TMEM100 was clearly lower in breast cancer tissues (P < 0.001) [Figure 1a]. Here, we verified TMEM100 expression in 40 breast cancer tissues collected from our hospital. The results were similar, the overall expression of TMEM100 in breast cancer tissues showed a downward trend (P < 0.01) [Figure 1b and c]. IHC results further showed that TMEM100 was downregulated in breast cancer tissues [Figure 1d].
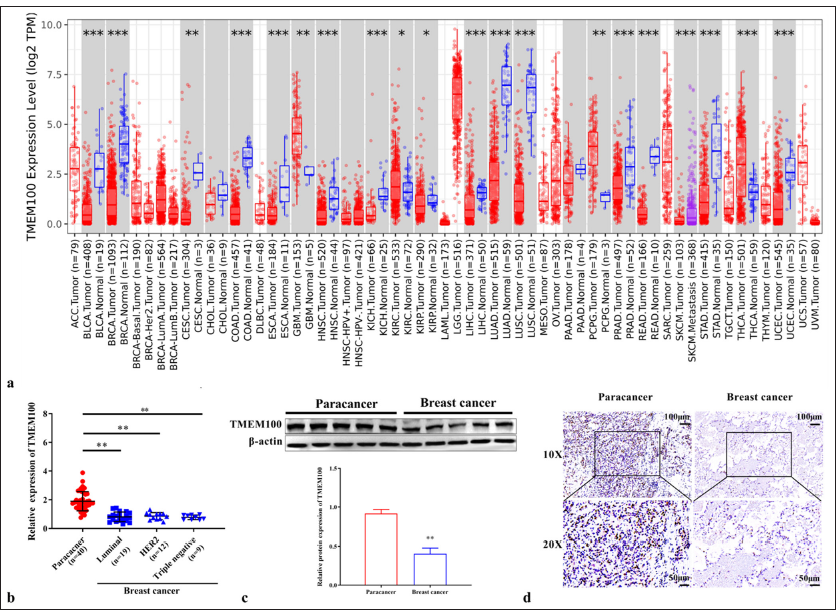
- Downregulation of TMEM100 in breast cancer tissues. (a) TMEM100 expression in different cancers in the TIME2.0 database. (b) RTqPCR results showing the mRNA expression of TMEM100 in breast cancer tissues (n=40). (c) Western blot results showing the protein expression of TMEM100 in breast cancer tissues (n=40). (d) IHC staining of TMEM100 protein expression in breast cancer tissues and paracancer tissues (n=5, 10×: 100 μm; 20×: 50 μm). ✶ P < 0.05, ✶✶ P < 0.01, ✶✶✶ P < 0.001. TMEM100: Transmembrane protein 100, RT-qPCR: Real-time quantity polymerase chain reaction, IHC staining: Immunohistochemical staining. ACC: Adrenocortical carcinoma, BLCA: Bladder Urothelial Carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and Neck squamous cell carcinoma, HPV: Human Papillomavirus, KICH: Kidney Chromophobe, KIRC: Kidney renal clear cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, TGCT: Testicular germ cell tumors, THCA: Thyroid carcinoma, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma, UCS: Uterine, UVM: Uveal melanoma, HER2: human epidermal growth factor receptor-2.
Prohibited the invasion and migration of breast cancer cells
Next, we found that TMEM100 expression was relatively lower in MCF-7 (P < 0.01), MDA-MB-231 (P < 0.01), T47D (P < 0.01), BT474 (P < 0.01), and SK-BR-3 cells (P < 0.01) than MCF-10A cells [Figure 2a]. Subsequently, functional-verification experiments were performed by transferring TMEM100 overexpressed sequences into MCF-7 and BT474 cells. The successful overexpression of TMEM100 in MCF-7 and BT474 cells was confirmed by RT-qPCR and Western blot (P < 0.01) [Figure 2b and c]. As shown in [Figure 2d], after transfection with TMEM100, the cell invasiveness decreased significantly (P < 0.01). Similarly, TMEM100 reduced the migration ability of breast cancer cells (P < 0.01) [Figure 2e].
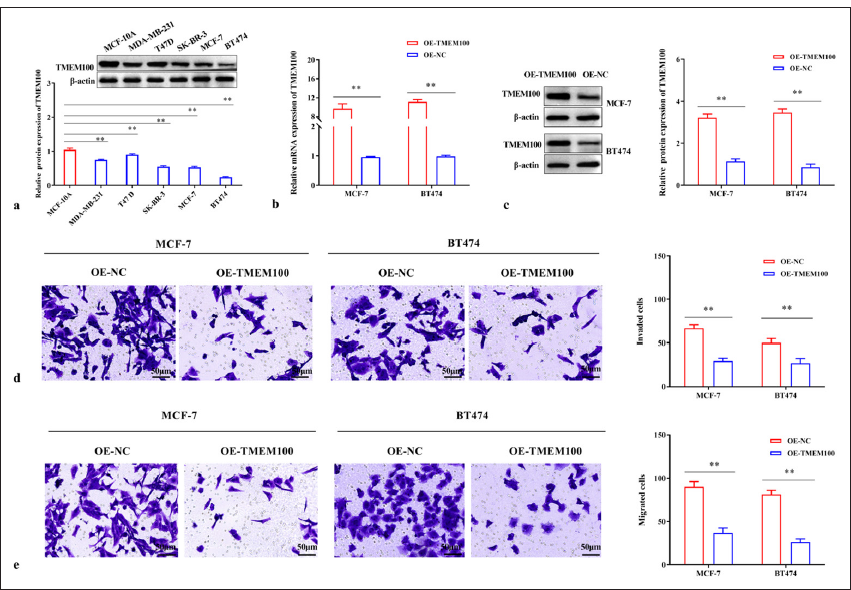
- TMEM100 prohibited the invasiveness and migration in breast cancer. (a) Relative protein expression of TMEM100. (b) RT-qPCR and (c) Western blot were used to detect the efficiency of TMEM100 overexpression. Transwell assay manifested that the overexpression of TMEM100 prohibited (d) cell invasiveness and (e) cell migration. ✶✶ P < 0.01, n = 3. TMEM100: Transmembrane protein 100, RT-qPCR: Real-time quantity polymerase chain reaction, OE-NC: Overexpressing negative control, OE-TMEM100: Overexpressing TMEM100.
TMEM100 prohibited the cell proliferation in breast cancer
After transfecting with TMEM100, the number of cell clones was dramatically reduced (P < 0.01) [Figure 3a]. In addition, the EdU-positive rate of TMEM100 transfected cells was decreased significantly (P < 0.01) [Figure 3b]. Therefore, our results suggested that TMEM100 prohibited the proliferative ability of breast cancer cells.
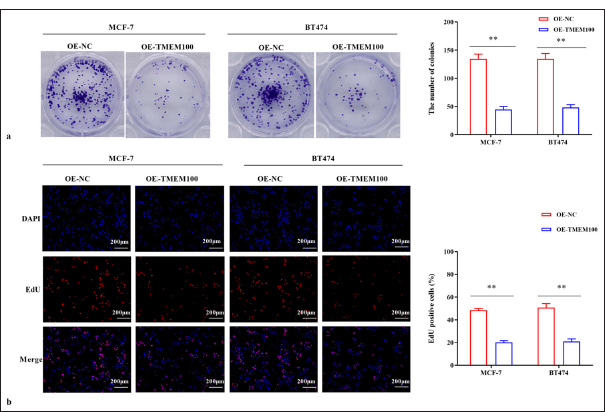
- TMEM100 prohibited the cell proliferation in breast cancer. (a) Cell colony-formation assay verified that TMEM100 overexpression decreased the number of cell clones. (b) EdU assay showed that TMEM100 overexpression decreased the EdU-positive cell rate. ✶✶ P < 0.01, n = 3. TMEM100: Transmembrane protein 100, EdU: 5-Ethynyl-2-deoxyuridine, OE-NC: Overexpressing negative control, DAPI: 4’,6-diamidino-2-phenylindole.
TMEM100 increased the chemosensitivity of breast cancer cells to docetaxel
Docetaxel was added to MCF-7 and BT474 cells with a concentration gradient. MTT results showed that docetaxel significantly hindered cell growth with increased docetaxel dose. After 24 h of treatment, the half-maximal inhibitory concentration (IC50) was 20.45 and 16.06 nM, respectively (P < 0.01) [Figure 4a and b]. Next, docetaxel at about half the dose of IC50 was used to treat MCF-7 cells (10 nM) and BT474 cells (8 nM). Notably, TMEM100 clearly enhanced docetaxel-induced decline in cell viability (P < 0.01) [Figure 4c] and (P < 0.05) [Figure 4d]. TMEM100 also increased the docetaxel-induced apoptosis of MCF-7 and BT474 cells (P < 0.01) [Figure 4e and f]. Together, our findings demonstrated that TMEM100 increased the chemosensitivity of breast cancer cells to docetaxel.
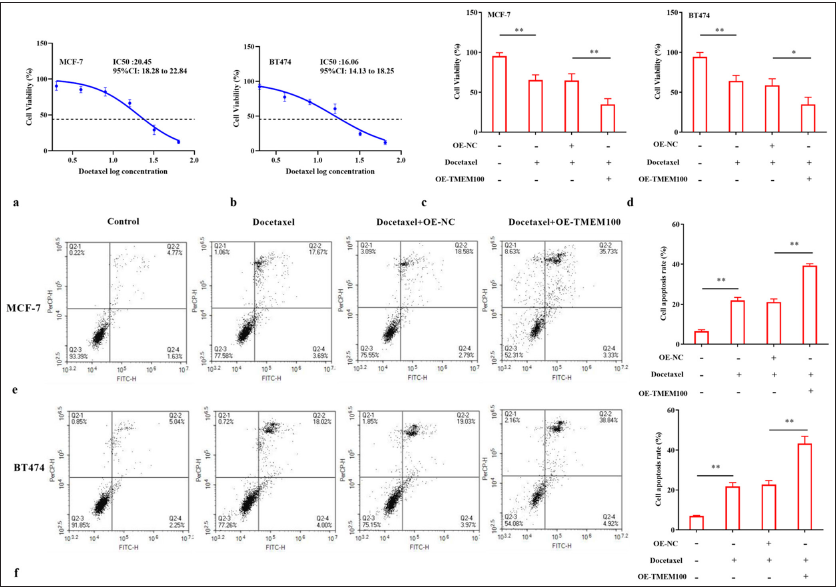
- TMEM100 increased the chemosensitivity of breast cancer cells to docetaxel. IC50 cytotoxicity assay confirmed the resistance of (a) MCF-7 cells and (b) BT474 cells to docetaxel. MTT assay showed that TMEM100 enhanced the sensitivity of (c) MCF-7 cells and (d) BT474 cells to docetaxel. TMEM100 notably increased the docetaxel-induced apoptosis of (e) MCF-7 cells and (f) BT474 cells. ✶ P < 0.05, ✶✶ P < 0.01, n = 3. TMEM100: Transmembrane protein 100, MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium OE-NC: Overexpressing negative control.
TMEM100 modulated the RAS/ERK pathway
TMEM100 reduced RAS expression and reduced the phosphorylation levels of p-ERK1/2 and p-MEK (P < 0.01) [Figure 5a and b]. ERK activator Tert-Butylhydroquinone (TBHQ) reversed the inhibition of TMEM100 on the RAS/ERK pathway (P < 0.01) [Figure 5c and d]. TBHQ reversed the inhibition of TMEM100 on cell invasiveness in MCF-7 cells (P < 0.01) and BT474 cells (P < 0.01) [Figure 6a]. Additionally, TBHQ reversed the inhibition of TMEM100 on cell migration in MCF-7 cells (P < 0.01) and BT474 cells (P < 0.05) [Figure 6b]. Furthermore, the inhibitory effect of TMEM100 on cell proliferation was reversed by TBHQ (P < 0.01) [Figure 6c-e]. Therefore, TMEM100 hindered the progression of breast cancer by blocking the RAS/ERK pathway.
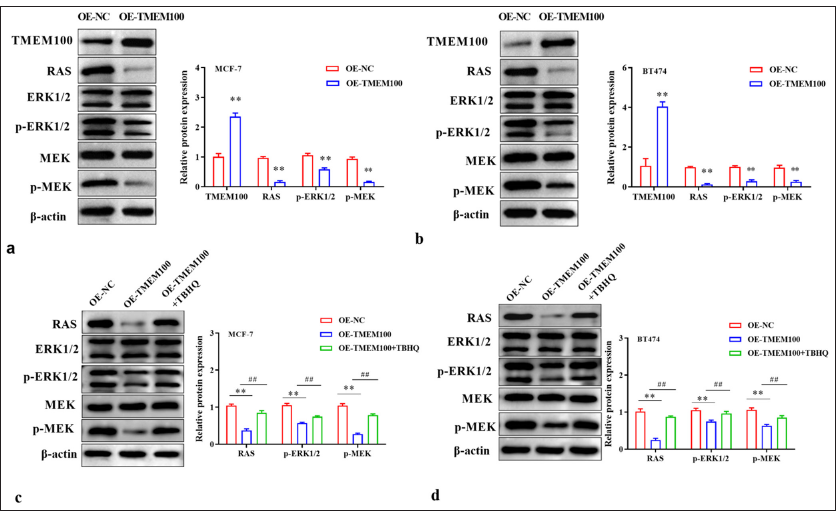
- TMEM100 modulated the RAS/ERK pathway. Effects of TMEM100 overexpression on RAS/ERK pathway-related protein expression in (a) MCF-7 cells and (b) BT474 cells were evaluated by Western blot assay. Effects of TMEM100 overexpression and TBHQ on RAS/ERK pathway-related protein expression in (c) MCF-7 cells and (d) BT474 cells were examined by Western blot assay. ## P < 0.01, versus OE-TMEM100 group; ✶✶ P < 0.01, versus OE-NC group, n = 3. TMEM100: Transmembrane protein 100, RAS/ERK: RAS-extracellular signal-regulated kinase, TBHQ: Tert-Butylhydroquinone OE-NC: Overexpressing negative control, OE-TMEM100: Overexpressing TMEM100.
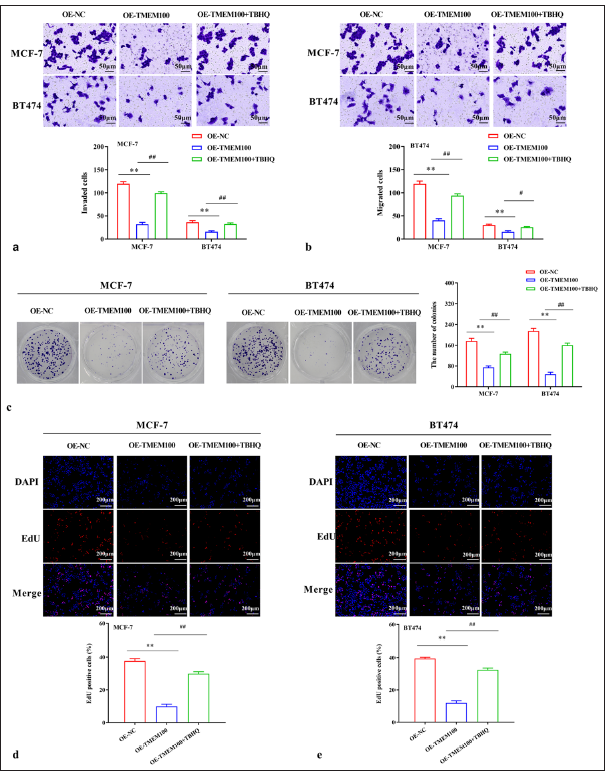
- Inhibitory effect of TMEM100 was related to the inactivation of the RAS/ERK pathway. (a) Transwell assay demonstrated that TBHQ reversed the decline in cell invasiveness caused by TMEM100. (b) Transwell assay manifested that TBHQ reversed the decline in cell migration caused by TMEM100. (c) Cell colony-formation assay showed that TBHQ reversed the decrease in the number of cell clones caused by TMEM100. (d, e) EdU assay exhibited that TBHQ reversed the decline in EdU-positive cell rate. # P < 0.05, ## P < 0.01, versus OE-TMEM100 group; ✶✶ P < 0.01, versus OE-NC group, n = 3. TMEM100: Transmembrane protein 100, RAS/ERK: RAS-extracellular signal-regulated kinase, TBHQ: Tert-Butylhydroquinone, OE-NC: Overexpressing negative control, DAPI: 4’,6-diamidino-2-phenylindole, EdU: 5-ethynyl-2′-deoxyuridine.
DISCUSSION
As a disease seriously endangers women’s health, the etiology of breast cancer is still uncertain. TMEM100 plays a particular role in various human cancers. Guo et al. found that the necrosis volume of glioma is positively correlated with TMEM100, whereas the volume of enhanced glioma is negatively correlated with it.[17] Furthermore, TMEM100 can effectively predict first progression, overall survival, and post-progression survival in gastric cancer.[18] In addition, TMEM100 reduces the malignancy of prostate cancer by hindering the proliferation and metastasis of malignant cells.[19] The role of TMEM100 in breast cancer is ambiguous so we want to explore whether the abnormal expression of TMEM100 can also affect the malignant process of breast cancer.
Consistent with the results presented by the TIME2.0 database, we found that TMEM100 decreased in breast cancer tissues and cells. Functional experiments revealed that TMEM100 overexpression clearly hindered the multiplication and metastasis of breast cancer cells. TMEM100 overexpression prohibited the invasion, migration, and proliferation activities of hepatocellular carcinoma.[20] The above research is basically consistent with our results. TMEM100 also reportedly inhibits the angiogenesis induction and migration in colorectal cancer.[21] Therefore, we can conclude that TMEM100 also acts as a cancer suppressor gene in breast cancer by impeding the growth and metastasis of cancer cells.
Docetaxel is a commonly used chemotherapy drug that promotes the growth of microtubules and blocks the depolymerization of microtubules. The outcome is effectively preventing the growth of tumor cells at G2/M stage.[22] Due to the resistance of patients, the therapeutic effect of chemotherapy is limited. Therefore, understanding the resistance mechanism of docetaxel in breast cancer and making corresponding clinical decisions can provide better treatment for patients. Here, TMEM100 enhanced the chemical sensitivity of breast cancer cells to docetaxel. Similarly, breast cancer cells carrying TMEM100 amplification are particularly sensitive to neoadjuvant chemotherapy (fluorouracil/epirubicin/cyclophosphamide and taxane) regimens.[23] Moreover, Zhuang et al. found that TMEM100 overexpression hinders the metastasis and restores the sensitivity of gastric cancer cells to cisplatin and 5-fluorouracil.[24]
Next, we investigated the signaling pathway involved in regulating TMEM100 during breast cancer cell progression. Phosphorylation activation of ERK1/2 leads to an abnormal increase in cell proliferation and migration, thereby exacerbating the malignancy of tumor cells.[25,26] We found that TMEM100 reduced RAS level and phosphorylation levels of p-ERK1/2 and p-MEK, and ERK agonists TBHQ neutralized the inhibition of TMEM100. Hence, it was concluded that TMEM100 hindered the malignant process of breast cancer by inhibiting RAS/ERK pathway.
Due to the small number of cases selected in this study and the high loss of follow-up rate, it is impossible to analyze the impact of TMEM100 on the prognosis. Future research can explore the interaction between TMEM100 expression and clinical outcomes in breast cancer patients, including survival, chemotherapy response, and disease recurrence. Moreover, subsequent trials should consider validation in clinically relevant breast cancer samples. In addition, subsequent bioinformatic analyses such as gene set enrichment analysis or pathway analysis should be considered to elucidate the functional role and biological processes of gene-expression profiles associated with TMEM100 in breast cancer. Furthermore, in follow-up trials, we shall consider determining whether TMEM100 modulates other signaling pathways in breast cancer.
SUMMARY
TMEM100 prohibited the cell growth and metastasis and enhanced the chemical sensitivity to docetaxel in breast cancer by hindering RAS/ERK pathway. We speculated that TMEM100 may become a high-potential target for breast cancer and play a pivotal role in future precision medicine for breast cancer patients.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
ABBREVIATIONS
TMEM100 – Transmembrane protein 100
RT-qPCR – Real-time quantitative polymerase chain reaction
EdU – 5-Ethynyl-2-deoxyuridine
FBS – Fetal bovine serum
IHC staining – Immunohistochemical staining
OE-NC – Overexpressing negative control
MTT – 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
RAS/ERK – RAS-extracellular signal-regulated kinase
TBHQ – Tert-Butylhydroquinone
FCM – Flow cytometry
AUTHOR CONTRIBUTIONS
XL and GQZ: Designed the research study; LZ: Performed the research; XL and LZ: Collected and analyzed the data. All authors have been involved in drafting the manuscript and all authors have been involved in revising it critically for important intellectual content. All authors give final approval of the version to be published. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This research was approved by the ethics committee of Zibo Central Hospital, approval No. 2024129, dated 2024. All experiments were performed in accordance with the ethical guidelines of Helsinki Declaration and with the informed consent of all patients.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-49.
- [CrossRef] [PubMed] [Google Scholar]
- Serum androgen dynamics in young women aged 18-40 treated with chemotherapy for breast cancer: An observational, multicentric, prospective study in France. Hum Fertil (Camb). 2024;27:2350758.
- [CrossRef] [PubMed] [Google Scholar]
- Immunotherapy for breast cancer treatment: Current evidence and therapeutic options. Endocr Metab Immune Disord Drug Targets. 2022;22:212-24.
- [CrossRef] [PubMed] [Google Scholar]
- Breast cancer: Current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51-64.
- [CrossRef] [PubMed] [Google Scholar]
- Adjuvant and neoadjuvant treatment of triple-negative breast cancer with chemotherapy. Cancer J. 2021;27:41-9.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanisms of docetaxel resistance in prostate cancer: The key role played by miRNAs. Biochim Biophys Acta Rev Cancer. 2021;1875:188481.
- [CrossRef] [PubMed] [Google Scholar]
- TMEM proteins in cancer: A review. Front Pharmacol. 2018;9:1345.
- [CrossRef] [PubMed] [Google Scholar]
- TMEM17 promotes malignant progression of breast cancer via AKT/GSK3β signaling. Cancer Manag Res. 2018;10:2419-28.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of TMEM178 as a potential prognostic biomarker and therapeutic target for breast cancer. Iran J Public Health. 2023;52:2427-39.
- [CrossRef] [PubMed] [Google Scholar]
- Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685-90.
- [CrossRef] [PubMed] [Google Scholar]
- The role of a lung vascular endothelium enriched gene TMEM100. Biomedicines. 2023;11:937.
- [CrossRef] [PubMed] [Google Scholar]
- Essential role for TMEM100 in vascular integrity but limited contributions to the pathogenesis of hereditary haemorrhagic telangiectasia. Cardiovasc Res. 2015;105:353-60.
- [CrossRef] [PubMed] [Google Scholar]
- TMEM100 is a key factor for specification of lymphatic endothelial progenitors. Angiogenesis. 2020;23:339-55.
- [CrossRef] [PubMed] [Google Scholar]
- TMEM100, a regulator of TRPV1-TRPA1 interaction, contributes to temporomandibular disorder pain. Front Mol Neurosci. 2023;16:1160206.
- [CrossRef] [PubMed] [Google Scholar]
- Low-expression of TMEM100 is associated with poor prognosis in non-small-cell lung cancer. Am J Transl Res. 2017;9:2567-78.
- [Google Scholar]
- Regulation of TMEM100 expression by epigenetic modification, effects on proliferation and invasion of esophageal squamous carcinoma. World J Clin Oncol. 2024;15:554-65.
- [CrossRef] [PubMed] [Google Scholar]
- The effect Of vascular related CeRNA genes and corresponding imaging biomarkers on survival in lower grade glioma. Ir J Med Sci. 2024;193:653-663.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of the diagnostic genes and immune cell infiltration characteristics of gastric cancer using bioinformatics analysis and machine learning. Front Genet. 2022;13:1067524.
- [CrossRef] [PubMed] [Google Scholar]
- GATA binding protein 5-mediated transcriptional activation of transmembrane protein 100 suppresses cell proliferation, migration and epithelial-to-mesenchymal transition in prostate cancer DU145 cells. Bioengineered. 2022;13:7972-83.
- [CrossRef] [PubMed] [Google Scholar]
- Novel roles of TMEM100: Inhibition metastasis and proliferation of hepatocellular carcinoma. Oncotarget. 2015;6:17379-90.
- [CrossRef] [PubMed] [Google Scholar]
- Transmembrane protein 100 inhibits the progression of colorectal cancer by promoting the ubiquitin/proteasome degradation of HIF-1α. Front Oncol. 2022;12:899385.
- [CrossRef] [PubMed] [Google Scholar]
- Exisulind in combination with docetaxel inhibits growth and metastasis of human lung cancer and prolongs survival in athymic nude rats with orthotopic lung tumors. Clin Cancer Res. 2002;8:904-12.
- [Google Scholar]
- Time series analysis of neoadjuvant chemotherapy and bevacizumab-treated breast carcinomas reveals a systemic shift in genomic aberrations. Genome Med. 2018;10:92.
- [CrossRef] [PubMed] [Google Scholar]
- TMEM100 expression suppresses metastasis and enhances sensitivity to chemotherapy in gastric cancer. Biol Chem. 2020;401:285-96.
- [CrossRef] [PubMed] [Google Scholar]
- Navigating the ERK1/2 MAPK cascade. Biomolecules. 2023;13:1555.
- [CrossRef] [PubMed] [Google Scholar]
- Peroxiredoxin 3 regulates breast cancer progression via ERK-mediated MMP-1 expression. Cancer Cell Int. 2024;24:59.
- [CrossRef] [PubMed] [Google Scholar]








