Translate this page into:
Jumonji domain-containing protein 6 promotes gastric cancer progression: Modulating immune evasion through autophagy and oxidative stress pathways

*Corresponding author: Di Na, Department of Surgical Oncology and General Surgery, The First Hospital of China Medical University, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors (China Medical University), Ministry of Education, Shenyang, Liaoning Province, China. zzz2023120828@163.com
-
Received: ,
Accepted: ,
How to cite this article: Zhang X, Na D. Jumonji domain-containing protein 6 promotes gastric cancer progression: Modulating immune evasion through autophagy and oxidative stress pathways. CytoJournal. 2025;22:6. doi: 10.25259/Cytojournal_230_2024
Abstract
Objective:
Immune response is crucial in the development of gastric cancer (GC), and Jumonji domain-containing protein 6 (JMJD6) plays an important role in mediating GC cell behavior. This study aims to elucidate the mechanisms through which JMJD6 affects autophagy and immune evasion in GC cells.
Material and Methods:
Immunocytochemistry was employed to assess JMJD6 and programmed death-ligand 1 (PD-L1) levels in gastric cancer cell line (MKN-45) and gastric epithelial cell line cells. MKN-45 cells with JMJD6 knockdown and overexpression were generated. The effect of JMJD6 on MKN-45 cells was evaluated using cell counting kit-8 assay, cellular fluorescence staining, and Transwell assays. Western blot analysis and immunofluorescence techniques were employed to investigate the regulation of autophagy by JMJD6. Reactive oxygen species (ROS) levels were evaluated by applying ROS fluorescence staining. Meanwhile, the protein and gene expression levels of molecules related to antioxidant stress responses were assessed through immunofluorescence assays and quantitative real-time polymerase chain reactions, respectively.
Results:
The expression levels of JMJD6 and PD-L1 were elevated in GC cells (P < 0.001). JMJD6 overexpression enhanced MKN-45 cell migration, invasion, and colony formation in vitro (P < 0.001). In MKN-45 cells, the epithelial-mesenchymal transition was promoted by JMJD6 upregulation but was notably inhibited by JMJD6 knockdown (P < 0.001). JMJD6 overexpression increased the expression levels of Sequestosome 1, Microtubule-associated protein 1A/1B-light chain 3 (LC3)II/LC3I, and PD-L1 in MKN-45 cells, and autophagy activation further elevated PD-L1 levels (P < 0.001). In addition, JMJD6 overexpression reduced ROS production and increased the expression of molecules related to antioxidant stress response, with the reverse effects observed on JMJD6 knockdown (P < 0.001).
Conclusion:
JMJD6 notably facilitates GC progression and immune evasion by modulating autophagy and oxidative stress pathways.
Keywords
Autophagy
Gastric cancer
Jumonji domain-containing protein 6
Oxidative stress
Programmed death-ligand 1
INTRODUCTION
Gastric cancer (GC) is a major global health concern due to its high incidence, fatality, and mortality rates in many countries.[1,2] Although recent advances have been made in the early screening and treatment of GC, its high recurrence rate and metastasis continue to pose a serious threat to patient prognosis.[3] Therefore, exploring the molecular mechanisms of GC and identifying new therapeutic targets remain current research priorities.
Jumonji domain-containing 6 (JMJD6) is a demethylase containing a Jumonji C domain and plays an important role in various cancers.[4,5] It regulates multiple biological processes, including transcription and the cell cycle, through its demethylation activity.[6,7] Although the role of JMJD6 in GC is particularly critical, its specific mechanisms remain incompletely understood.
Programmed death-ligand 1 (PD-L1) is a key immune system-related checkpoint compound that is widely expressed on the surfaces of various cancer cells.[8,9] By binding to the programmed death-1 (PD-1) receptor, PD-L1 inhibits T-cell responses, promoting the evasion of immune surveillance by tumor cells.[10,11] In recent years, the immune evasion mechanism of PD-L1 has become a major research focus in cancer immunotherapy.[12,13] Evidence has pointed to the importance of PD-L1 in GC and its potential as a target for therapy.[14] Given its central role in modulating the immune response, PD-L1 has gained attention as a promising target for immunotherapy in GC. Inhibiting tumor immunity with PD-1/PD-L1 inhibitors has shown potential in enhancing T-cell-mediated antitumor immunity, improving clinical outcomes, and offering a new therapeutic strategy for patients with advanced or refractory GC. Consequently, PD-L1 is not only a biomarker for predicting response to immunotherapy, but it is also an emerging target for developing novel treatment strategies aimed at overcoming immune resistance in GC.
Autophagy is a crucial mechanism by which cells respond to internal and external environmental stressors, allowing for the removal of damaged organelles and proteins to maintain cellular homeostasis.[15,16] In cancer, autophagy can either promote tumor cell survival or lead to tumor cell death.[17] Recent studies have highlighted its involvement in cancer cell survival and invasiveness, with emerging evidence suggesting that JMJD6 influences these processes through the regulation of autophagy, a cellular process essential for maintaining cellular homeostasis by degrading and recycling damaged organelles, proteins, and other cellular components.[18,19] However, the specific approach by which JMJD6 regulates autophagy and its effect on GC development remains to be fully elucidated.
We focus on exploring the involvement and underlying mechanisms of JMJD6 in GC to address the above knowledge gaps. We systematically examine the function of JMJD6 in GC cell reproduction, migration, and invasion using cell experiments. In addition, we explore how JMJD6 regulates PD-L1 expression through autophagy and the nuclear factor erythroid 2-related factor 2 (Nrf2) pathway. Our studies will contribute to understanding the oncogenic role of JMJD6 in GC.
MATERIAL AND METHODS
Cell cultures
The human gastric adenocarcinoma cell line gastric cancer cell line (MKN-45) (iCell-h345) and gastric epithelial tissue cell line (GES-1) (iCell-h062) were sourced from Cellverse Co., Ltd. (Shanghai, China). MKN-45 cells were cultured in their designated medium (iCell-h345-001b, Cellverse Co., Ltd., Shanghai, China), whereas GES-1 cells were cultured in medium (iCell-h062-001b, Cellverse Co., Ltd., Shanghai, China) specific for GES-1. All cells were maintained in a 37°C incubator (CB160, Binder, Tuttlingen, Baden-Württemberg, Germany) with 5% carbon dioxide (CO2). The cell lines used in this study were authenticated through Short Tandem Repeat profiling and were negative for mycoplasma contamination.
Cell transfection
First, MKN-45 cells were cultured in a six-well plate until they reached 60% confluence. Next, the short hairpin RNA (shRNA) negative control and ShRNA (JMJD6) (5 µg for a six-well plate) were mixed with Optimized Minimum Essential Medium and added with a transfection reagent (Lipofectamine 3000) (L3000150, Invitrogen, Waltham, Massachusetts, the USA). The mixture was incubated at 25°C for 15 min to form transfection complexes. The transfection complexes for the JMJDA overexpression plasmid were prepared in a similar manner. Each complex was added to the cells separately, gently swirled to mix, and incubated in an incubator (CB160, Binder, Tuttlingen, Baden-Württemberg, Germany) at 37°C and 5% CO2 for 4 h. Afterward, the medium was replaced with fresh culture medium. Incubation was continued for 48 h. Subsequently, transfection efficiency was assessed using quantitative reverse transcription polymerase chain reaction (qRT-PCR).
ShRNA-negative control (NC) interference sequences: TTCTCCGAACGTGTCACGT; ShRNAJMJD6 (Sh-JMJD6)-1 NC interference sequences: GCACAACTACTACGAGAGCTT; Sh-JMJD6-2 NC interference sequences: CGAAGCTATTACCTGGTTTAA;
Sh-JMJD6-3 NC interference sequences: ATGGACTCT GGAGCGCCTAAA; JMJDA overexpression interference sequences: GGAGCGGTATGAAAGACCTT ACAAGCCCGTGGTTTTGTTGAATGCG CAAGAGGGCTGGTCTGCGCAGGAGAA ATGGACTCTGGAGCGCCTAAAAAGGAA ATATCGGAACCAGAAGTTCAAGTGTGGT GAGGATAACGATGGCTACTCAGTGAAGAT GAAGATGAAATACTACATCGAGTACATGG AGAGCACTCGAGATGATAGTCCCCTTTACA TCTTTGACAGCAGCTATGGTGAACACCCT AAAAGAAGGAAACTTTTGGAAGACTACAA GGTGCCAAAGTTTTTCACTGATGACCTTT TCCAGTATGCTGGGGAGAAGCGCAGGCC CCCTTACAGGTGGTTTGTGATGGGGCCACC ACGCTCCGGAACTGGGATTCACATCGACC CTCTGGGAACCAGTGCCTGGAATGCCTTAGT TCAGGGCCACAAGCGCTGGTGCCTGTTTCCTAC CAGCACT.
Immunocytochemistry
First, MKN-45 cells were cultured to the logarithmic phase, fixed for 10 min, and then washed with phosphate-buffered saline (PBS). Next, the cells were permeabilized for 10 min and washed with PBS. Subsequently, non-specific binding was blocked through incubation with 5% Bovine Serum Albumin (BSA) for 1 h. Following this procedure, the cells were incubated with the specific primary antibodies JMJD6 (YP-Ab-12949, UpingBio, Hangzhou, China), PD-L1 (ab205921), Epithelial cadherin (E-cadherin) (ab314063), and Nrf2 (ab62352) at 4°C overnight; washed with PBS; incubated with a SAlexa Fluor 568–labeled secondary antibody (goat anti-rabbit Immunoglobulin G [IgG]) (K1034G-AF568, Solarbio, Beijing, China) or Alexa Fluor® 488–labeled secondary antibody (goat anti-mouse IgG) (ab150113) for 1 h; and washed again with PBS. SAlexa Fluor 568 and Alexa Fluor® were presenting red fluorescence. Stain the cell nuclei with 4’,6-diamidino-2-phenylindole (DAPI) (C1002, Beyotime, Shanghai, China) for 10 min. Next, the cells were washed with PBS. Finally, the cells were mounted in an antifade mounting medium, and images were observed and captured with a fluorescence microscope (Axio Observer 7, Zeiss, Heidenheim, Germany). ImageJ software (version 1.5f, NIH, Bethesda, Maryland, the USA) was employed for the quantitative analysis of the images. The primary antibodies (PD-L1, E-cadherin, and Nrf2) and Alexa Fluor® 488–labeled secondary antibody were purchased from Abcam Biotechnology Inc. (Abcam) (Cambridge, the UK). The antibody dilution ratio was 1:1000.
qRT-PCR
First, total RNA was extracted from cell samples using a kit (R0017S, Beyotime, Shanghai, China), and quantification and quality assessment were performed. Next, the extracted RNA was reverse-transcribed into cDNA by employing a kit (D7168S, Beyotime, Shanghai, China). Subsequently, the quantitative polymerase chain reaction (qPCR) mixture was prepared by combining the cDNA with qPCR reagents, primers, and fluorescent probes. The mixture was distributed into a 96-well plate, and amplification was performed with a reverse transcription polymerase chain reaction machine (LightCycler 96, Roche, Basel, Switzerland). Fluorescence detection was conducted to obtain Ct values (threshold cycle numbers), which were analyzed using the 2−ΔΔCt method for quantification. GAPDH served as the reference gene. The primer sequences utilized in this study are provided in Table 1.
| Primer name | Primer sequences (5'–3') |
|---|---|
| JMJD6-F | TCCACAGGGATAGCTTCCGA |
| JMJD6-R | AGCTTACGCTGAAAGCACCT |
| NQO1-F | TGCTTACACTTACGCTGCCAT |
| NQO1-R | CCAGTGGTGATGGAAAGCAC |
| HO1-F | GTGCCACCAAGTTCAAGCAG |
| HO1-R | CACGCATGGCTCAAAAACCA |
| GAPDH-F | GTGGATATTGTTGCCATCAATGACC |
| GAPDH-R | GCCCCAGCCTTCTTCATGGTGGT |
JMJD6: Jumonji domain-containing protein 6, NQO1: NAD (P) H quinone dehydrogenase 1, HO1: Heme oxygenase 1, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Western blot analysis
First, cell samples were processed with lysis buffer (P0013B, Beyotime, Shanghai, China) to release proteins, and centrifugation was performed to remove cell debris. The resulting supernatant was collected for protein quantification by a BCA kit (P0009, Beyotime, Shanghai, China). Next, proteins were separated through sodium dodecyl sulfate polyacrylamide gel electrophoresis. The proteins were then transferred from the gel to a polyvinylidene fluoride membrane (FFP19, Beyotime, Shanghai, China). The membrane was blocked with 5% BSA (P0007, Beyotime, Shanghai, China) to prevent nonspecific binding. After this step, the membrane was incubated with the specific primary antibodies E-cadherin (ab314063), Neural cadherin (N-cadherin) (ab76011), Snail family transcriptional repressor 1 (Snail) (ab216347), Twist family BHLH transcription factor 1 (Twist) (ab175430), Sequestosome 1 (p62) (ab207305), Microtubule-associated protein 1A/1Blight chain 3 (LC3) (ab51520), PD-L1 (ab213524), and GAPDH (ab9485) overnight at 4°C; washed with PBS; incubated with a secondary antibody (ab6721 and ab6728) conjugated with Horseradish Peroxidase (HRP); and washed again. Signals were developed using Enhanced Chemiluminescence (P0018S, Abcam, Cambridge, the UK) with HRP and detected with a gel imaging system (Alliance 9.7, Uvitec, Cambridge, the UK). ImageJ software (version 1.5f, NIH, Bethesda, Maryland, the USA) was utilized for quantitative image evaluation. All primary antibodies and the secondary antibody were purchased from Abcam (Cambridge, the UK). The antibody dilution ratio was 1:1000.
Cell counting kit-8 (CCK-8) assay
The transfected MKN-45 cells were seeded into a 96-well plate, and 100 µL of a cell solution (1 × 106 cells/mL) was added per well. Incubation was continued for an additional 48 h. After treatment, 10 µL of CCK-8 reagent (C0037) was added to each well, allowing the CCK-8 reagent to react with the dehydrogenases in the cells and produce an orange-yellow formazan product. Absorbance was measured at 450 nm using a microplate reader (EnVision™, PerkinElmer, Waltham, Massachusetts, the USA). The CCK-8 reagent was bought from Beyotime (Shanghai, China). Cell viability (%) = (OD [treated group] − OD [blank group])/(OD [control group] − OD [blank group]) × 100%.
5-ethynyl-2'-deoxyuridine (EdU) staining
First, MKN-45 cells were seeded into a six-well plate for 24 h, added with 10 µM EdU (C0071L, Beyotime, Shanghai, China), and incubated for another 1 h. Afterward, the cells were fixed for 10 min and washed with PBS. Next, the cells were permeabilized for 10 min and washed again with PBS. Subsequently, the cells were incubated with Click-iT reaction mixture and incubated in the dark for 30 min to allow EdU to bind to fluorescently labeled azide (green) cells. The cells were then washed with PBS. Subsequently, cell nuclei were stained with DAPI, and the cells were washed again. Finally, images of EdU-stained cells were recorded under a fluorescence microscope (Axio Observer 7, Zeiss, Heidenheim, Germany), and EdU-positive cells were quantified by employing ImageJ software (version 1.5 f, National Institutes of Health, Bethesda, Maryland, the USA) to assess cell proliferation.
Transwell assay
Cell migration
MKN-45 cells were cultured in a serum-free medium for 24 h using Transwell chambers (FTW001, Beyotime, Shanghai, China) without Matrigel coating. A total of 1 × 105 MKN-45 cells were suspended and seeded in the upper chamber. Medium containing 10% fetal bovine serum (FBS) was added to the lower chamber as a chemoattractant. The cells were incubated for 1 day. Once incubation was completed, non-migrated cells were gently removed from the upper chamber, rinsed, and fixed for 10 min. The migrated cells were stained with 0.1% crystal violet (A1251, Sigma, Merck, Darmstadt, Germany) for 10 min then washed. The cells were assessed and counted using a light microscope (BX43, Olympus, Tokyo, Japan).
Cell invasion
Matrigel (C0376, Beyotime, Shanghai, China) was applied onto the upper chambers of a Transwell plate (FTW001, Beyotime, Shanghai, China) and incubated for 30 min to 1 h to allow solidification. A total of 1 × 105 MKN-45 cells were suspended in a serum-free medium and seeded in the upper chamber containing Matrigel. Medium containing 10% FBS was added to the lower chamber. Non-invading cells were removed and washed with PBS, then fixed with 4% paraformaldehyde for 10 min. The invading cells were fixed with 0.1% crystal violet (A1251, Sigma, Merck, Darmstadt, Germany) for 10 min, then washed with PBS. The cells were assessed and counted using a light microscope (BX43, Olympus, Tokyo, Japan).
Fluorescence staining of reactive oxygen species (ROS)
Transfected MKN-45 cells were seeded and incubated until they reached 70% confluence. Depending on the experimental design, the cells were treated with specific reagents or conditions to induce ROS production and incubated for 30 min to 2 h. Next, the cells were added with 10 µM fluorescent dichloro-dihydro-fluorescein diacetate (S0035S, Beyotime, Shanghai, China) and incubated in the dark for 30 min. After incubation, the cells were washed twice with PBS to remove excess dye. If needed, the cells were fixed for 10 min then washed. DAPI was used to stain the cell nuclei for visualization during imaging. Finally, the ROS-stained cells were observed under a fluorescence microscope (Axio Observer 7, Zeiss, Heidenheim, Germany). ImageJ software (version 1.5f, NIH, Bethesda, Maryland, the USA) was employed for quantitative image analysis
Statistical analysis
Statistical analysis was performed on the experimental results using GraphPad Prism software (version 9.0, GraphPad Software, Inc., San Diego, California, the USA). Data were presented as mean ± standard deviation. The t-test was employed for comparisons between groups. One-way ANOVA was applied to compare data among multiple groups and was followed by Tukey’s post hoc test. Herein, P < 0.05 was considered statistically significant.
RESULTS
High expression of JMJD6 and PD-L1 in GC
We validated the upregulation of JMJD6 and PD-L1 in GC cells through immunohistochemistry. The results in Figures 1a-d demonstrate that the expression levels of JMJD6 and PD-L1 were significantly elevated in MKN-45 cells relative to GES-1 cells (P < 0.001).
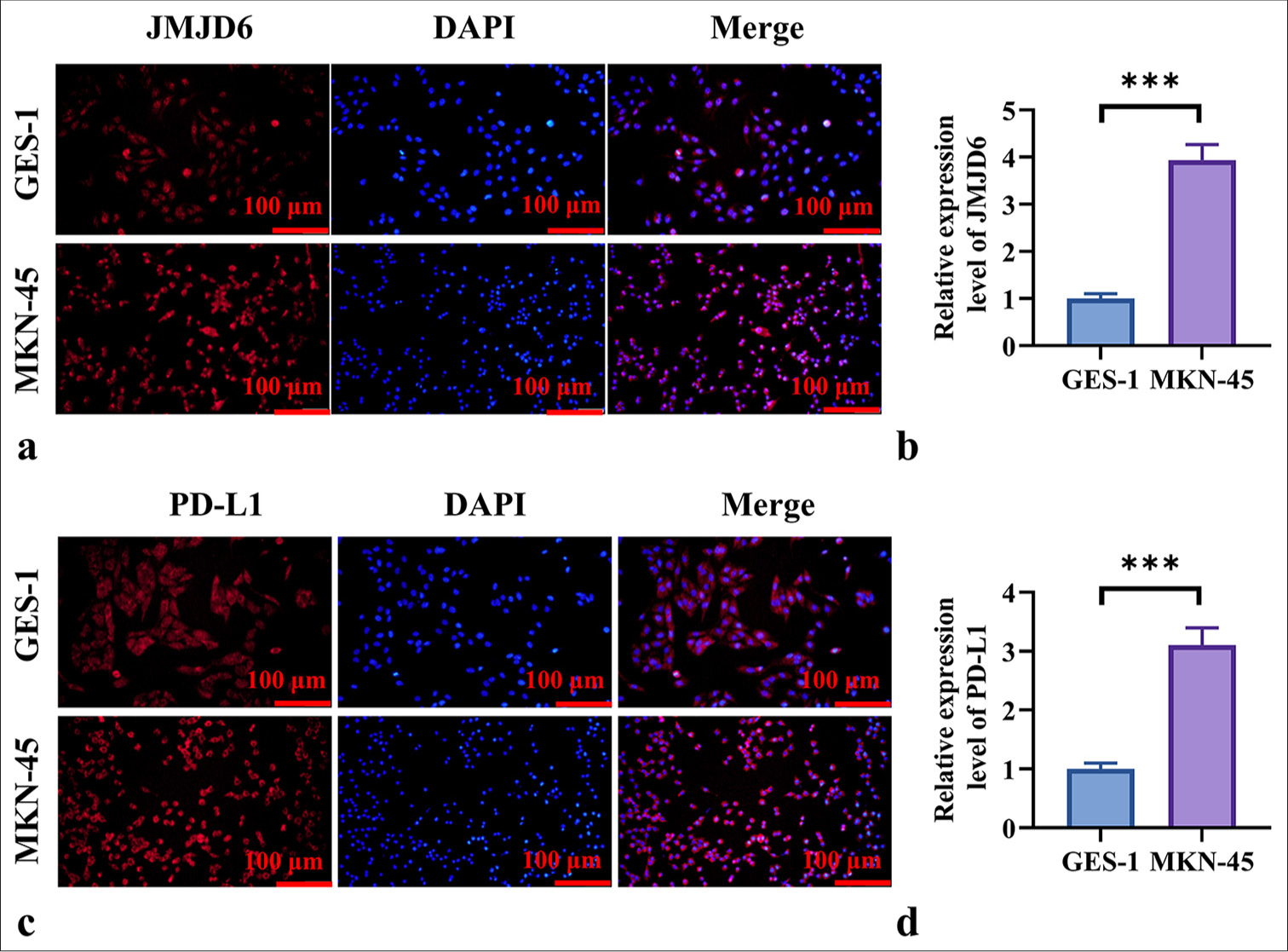
- High Expression of JMJD6 and PD-L1 in GC. (a and b) Immunohistochemistry staining results of JMJD6 in gastric epithelial cells (GES-1) and GC cells (MKN-45). (c and d) Immunohistochemistry staining results of PD-L1 in GES-1 and MKN-45 cells. n = 6. ✶✶✶P < 0.001. JMJD6: Jumonji domain–containing protein 6, GES-1: Gastric epithelial cell line, MKN-45: Gastric cancer cell line, GC: Gastric cancer, PD-L1: programmed death-ligand 1.
JMJD6 promotes the growth, migration, and invasion of GC cells
Next, we investigated the effects of JMJD6 on GC cells. ShRNA (JMJD6) and JMJD6 overexpression plasmids were transfected into MKN-45 cells, and the transfection efficiency of JMJD6 knockdown or overexpression was verified using qRT-PCR [Figure 2a]. On the basis of the results shown in Figure 2a, we selected Sh-JMJD6-1 as the transfection sequence for JMJD6 knockdown. We found that in the CCK-8 assay and EdU staining experiments, JMJD6 overexpression notably promoted MKN-45 cell proliferation (P < 0.001), whereas JMJD6 knockdown markedly inhibited GC cell proliferation (P < 0.001) [Figure 2b-d]. The Transwell assay further demonstrated that the number of migrated and invaded cells was notably higher in the JMJD6 overexpression group than in the negative control group (P < 0.001), whereas migration and invasion abilities notably reduced after JMJD6 knockdown (P < 0.001) [Figure 2e-h].
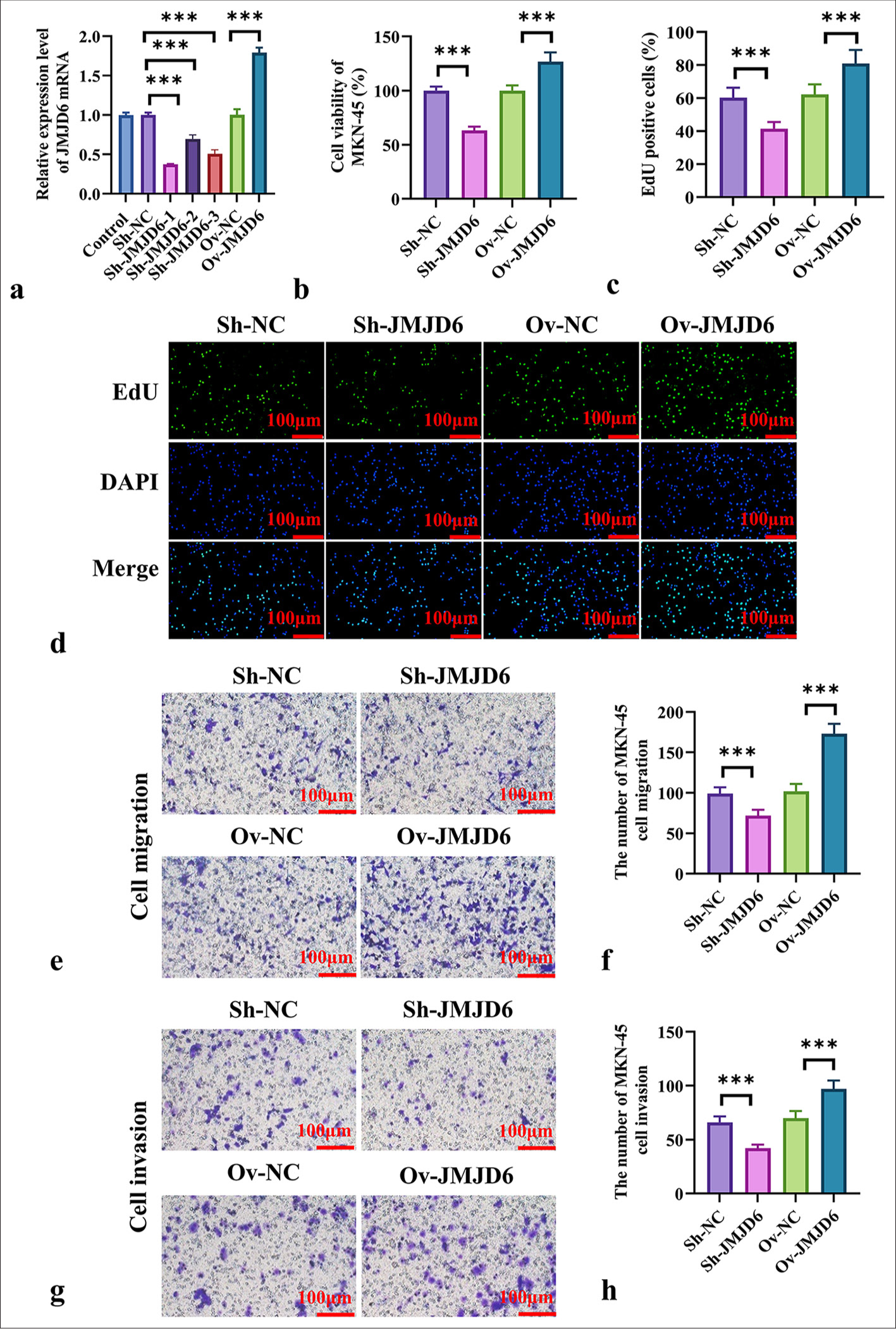
- JMJD6 promotes the proliferation, migration, and invasion of GC cells. (a) qRT-PCR validation of expression efficiency after JMJD6 knockdown and overexpression. (b) CCK-8 assay was used to assess the proliferation ability of MKN-45 cells after JMJD6 knockdown or overexpression. (c and d) EdU staining analysis of the number of EdU-positive cells following JMJD6 knockdown or overexpression. (e and f) Transwell assay was conducted to measure the migration capability of MKN-45 cells with different levels of JMJD6 expression. (g and h) Transwell assay was performed to assess the invasion ability of MKN-45 cells with different levels of JMJD6 expression. n = 6. ✶✶✶P < 0.001. JMJD6: Jumonji domain–containing protein 6, MKN-45: GC cell line, EdU: 5-ethynyl-2'-deoxyuridine, Sh-NC: ShRNA-negative control, ShJMJD6: ShRNA-JMJD6, Ov-NC: Overexpression-negative control, Ov-JMJD6: OverexpressionJMJD6, qRT-PCR: Quantitative reverse transcription polymerase chain reaction, CCK-8: Cell counting kit-8, GC: Gastric cancer, DAPI: 4’,6-diamidino-2-phenylindole
JMJD6 promotes epithelial-mesenchymal transition (EMT) in GC cells
We next assessed the changes in the expression levels of EMT-related markers following alterations in JMJD6 expression. Western blot analysis showed significant increases in N-cadherin, Snail, and Twist expression levels after JMJD6 overexpression (P < 0.001) [Figure 3a-e]. Conversely, JMJD6 knockdown led to a significant increase in E-cadherin protein levels (P < 0.001) [Figure 3a-e]. In addition, the immunofluorescence analysis of E-cadherin expression revealed that JMJD6 downregulation resulted in a significant increase in E-cadherin expression (P < 0.001) [Figure 3f and g], whereas JMJD6 upregulation led to a significant reduction in E-cadherin expression (P < 0.001) [Figure 3f and g].
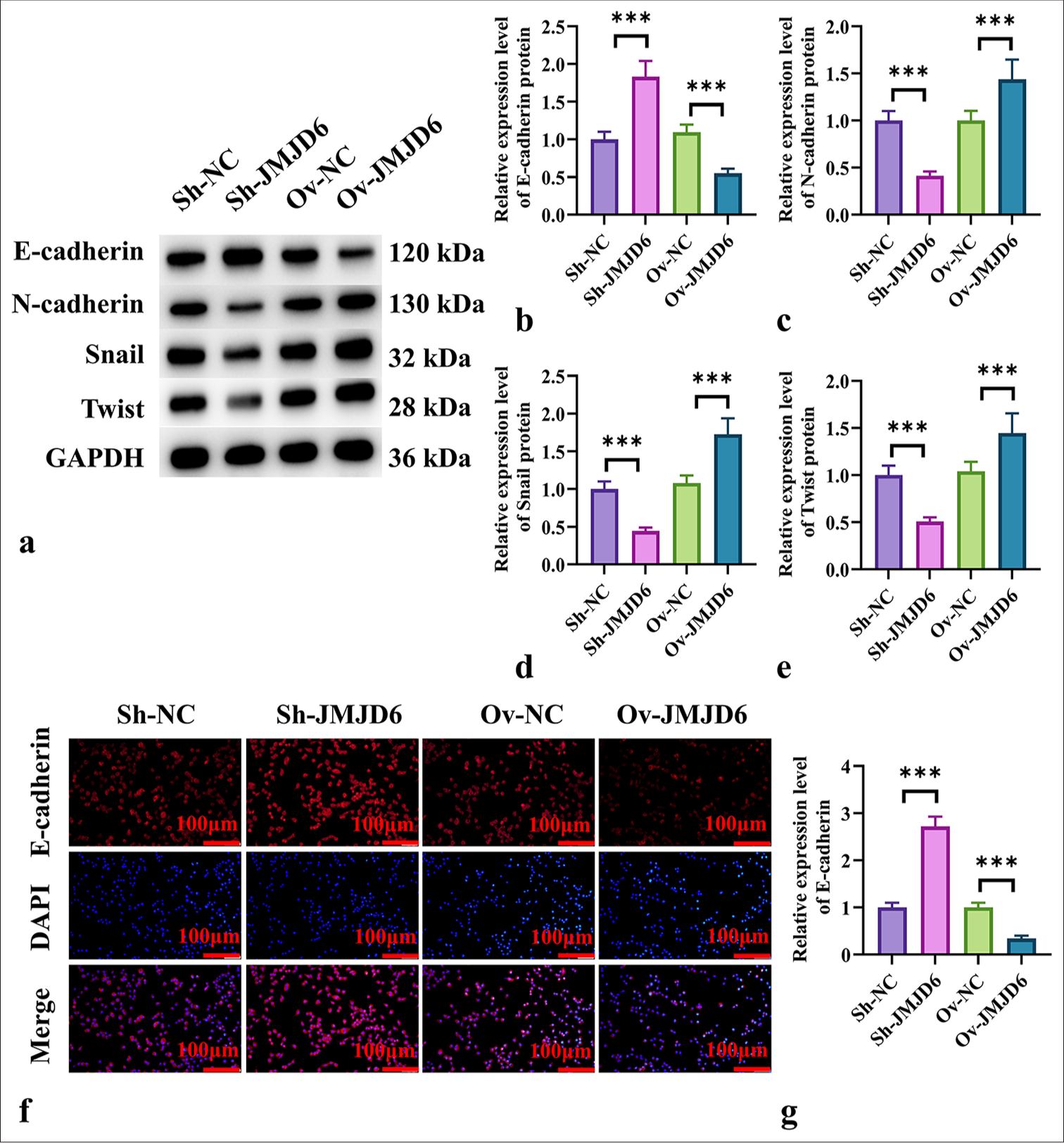
- JMJD6 Promotes EMT in GC Cells. (a-e) Western blot analysis of the EMT-related proteins E-cadherin, N-cadherin, Snail, and Twist following JMJD6 knockdown or overexpression. (f and g) Immunofluorescence analysis of E-cadherin expression in MKN-45 cells with different levels of JMJD6 expression. n = 6. ✶✶✶P < 0.001. JMJD6: Jumonji domain–containing protein 6, Sh-NC: ShRNA-negative control, Sh-JMJD6: ShRNA-JMJD6, Ov-NC: Overexpression-negative control, Ov-JMJD6: Overexpression-JMJD6, E-cadherin: Epithelial cadherin, N-cadherin: Neural cadherin, Snail: Snail family transcriptional repressor 1, Twist: Twist family BHLH transcription factor 1, GAPDH: glyceraldehyde-3-phosphate dehydrogenase, DAPI: 4',6-diamidino-2-phenylindole, GC: Gastric cancer, EMT: Epithelial–mesenchymal transition, MKN-45: Gastric cancer cell line.
JMJD6 increases PD-L1 expression in GC cells by activating autophagy
We hypothesized that JMJD6 and PD-L1 expression levels are positively correlated in GC. Immunofluorescence analysis revealed that PD-L1 expression in MKN-45 cells significantly increased on JMJD6 overexpression but significantly decreased following JMJD6 knockdown (P < 0.001) [Figure 4a and b]. We assessed the expression levels of autophagy-related proteins in MKN-45 cells after JMJD6 overexpression or knockdown to explore the relationship between JMJD6 and autophagy. The results shown in Figure 4c-f indicate that in JMJD6-overexpressing MKN-45 cells, LC3II/LC3I and p62 protein levels significantly elevated, paralleling the increase in PD-L1 protein expression (P < 0.001). Conversely, JMJD6 knockdown resulted in a significant reduction in LC3II/LC3I, p62, and PD-L1 protein levels (P < 0.001). This finding suggests that JMJD6 overexpression activates autophagy in MKN-45 cells.
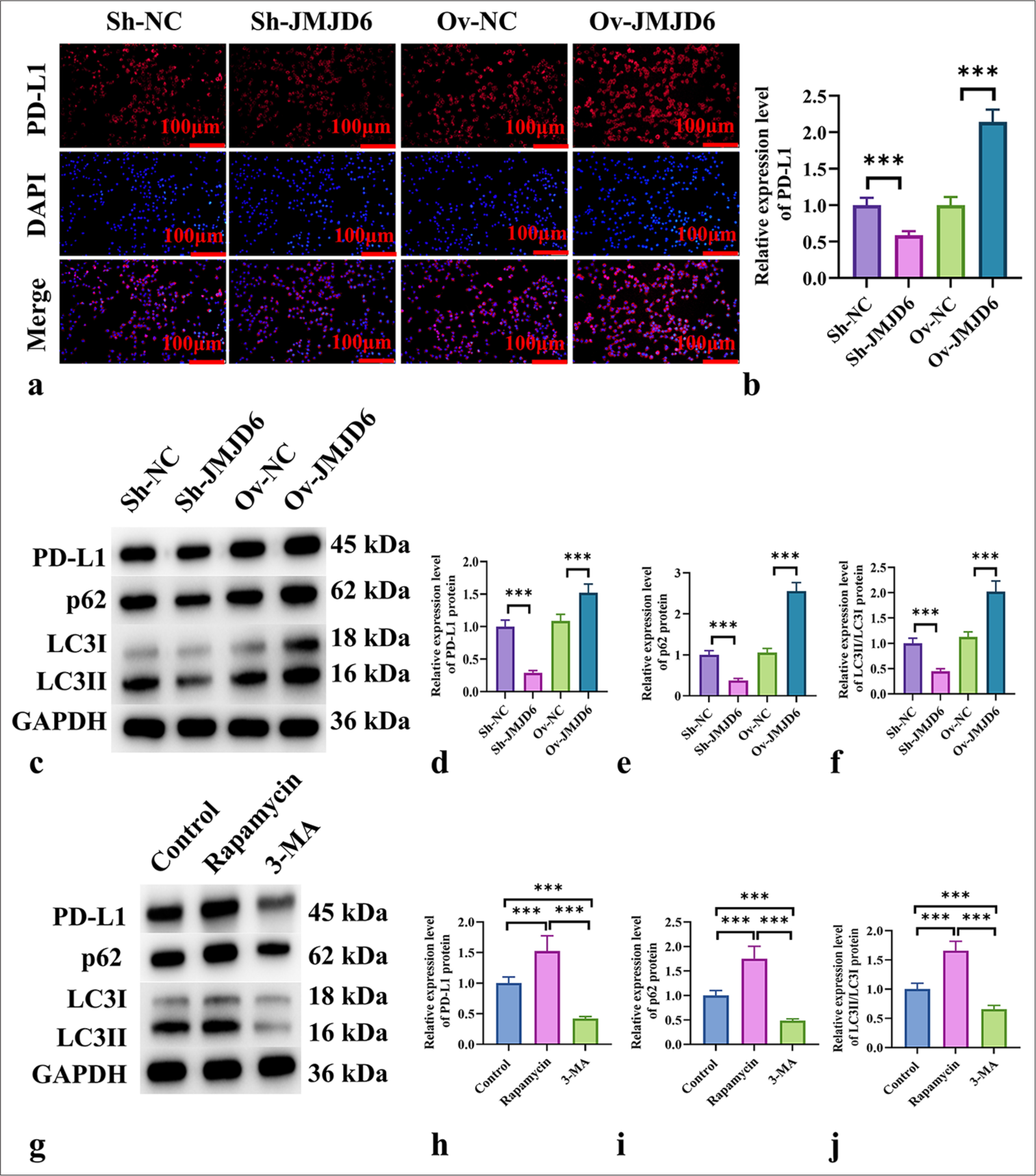
- JMJD6 increases PD-L1 expression in GC cells by activating autophagy. (a and b) Immunofluorescence analysis of PD-L1 expression in MKN-45 cells with different levels of JMJD6 expression. (c-f) Western blot analysis of PD-L1 and the autophagy-related proteins LC3II, LC3I, and p62. (g-j) Western blot analysis of PD-L1 and the autophagy-related proteins LC3II, LC3I, and p62 in MKN-45 cells treated with starvation, rapamycin, and 3-MA for 24 h. n = 6. ✶✶✶P < 0.001. JMJD6: Jumonji domain–containing protein 6, PD-L1: Programmed death-ligand 1, Sh-NC: ShRNA-negative control, Sh-JMJD6: ShRNA-JMJD6, Ov-NC: Overexpression-negative control, Ov-JMJD6: Overexpression-JMJD6, p62: Sequestosome 1, LC3: Microtubule-associated protein 1A/1B-light chain 3, 3-MA: 3-methyladenine, GC: Gastric cancer, MKN-45: Gastric cancer cell line, GAPDH: glyceraldehyde-3-phosphate dehydrogenase
We used starvation and rapamycin to activate autophagy and 3-methyladenine (3-MA) and bafilomycin A1 to block autophagy to confirm whether the increase in PD-L1 is due to autophagy activation. The results presented in Figure 4g-j demonstrate that autophagy activation led to significant increases in LC3II/LC3I, p62, and PD-L1 protein levels (P < 0.001), whereas autophagy inhibition resulted in significant reductions in these proteins (P < 0.001).
JMJD6 activates the Nrf2 signaling pathway during autophagy in GC cells
Next, we used immunofluorescence to assess the fluorescence intensity of ROS in GC cells with JMJD6 knockdown or overexpression. Figure 5a and b show that ROS fluorescence intensity significantly increased in GC cells with JMJD6 knockdown but significantly reduced with JMJD6 overexpression (P < 0.001). We also examined the effect of JMJD6 on Nrf2 expression levels in MKN-45 cells. We found that Nrf2 expression levels significantly increased in cells overexpressing JMJD6, whereas JMJD6 knockdown markedly reduced Nrf2 levels (P < 0.001) [Figure 5c and d]. In addition, the qRT-PCR analysis of NAD(P)H quinone dehydrogenase 1 (NQO1) and Heme oxygenase 1 (HO1) expression levels revealed that the mRNA levels of NQO1 and HO1 notably reduced in MKN-45 cells with JMJD6 knockdown, whereas they significantly increased with JMJD6 overexpression (P < 0.001) [Figure 5e and f].
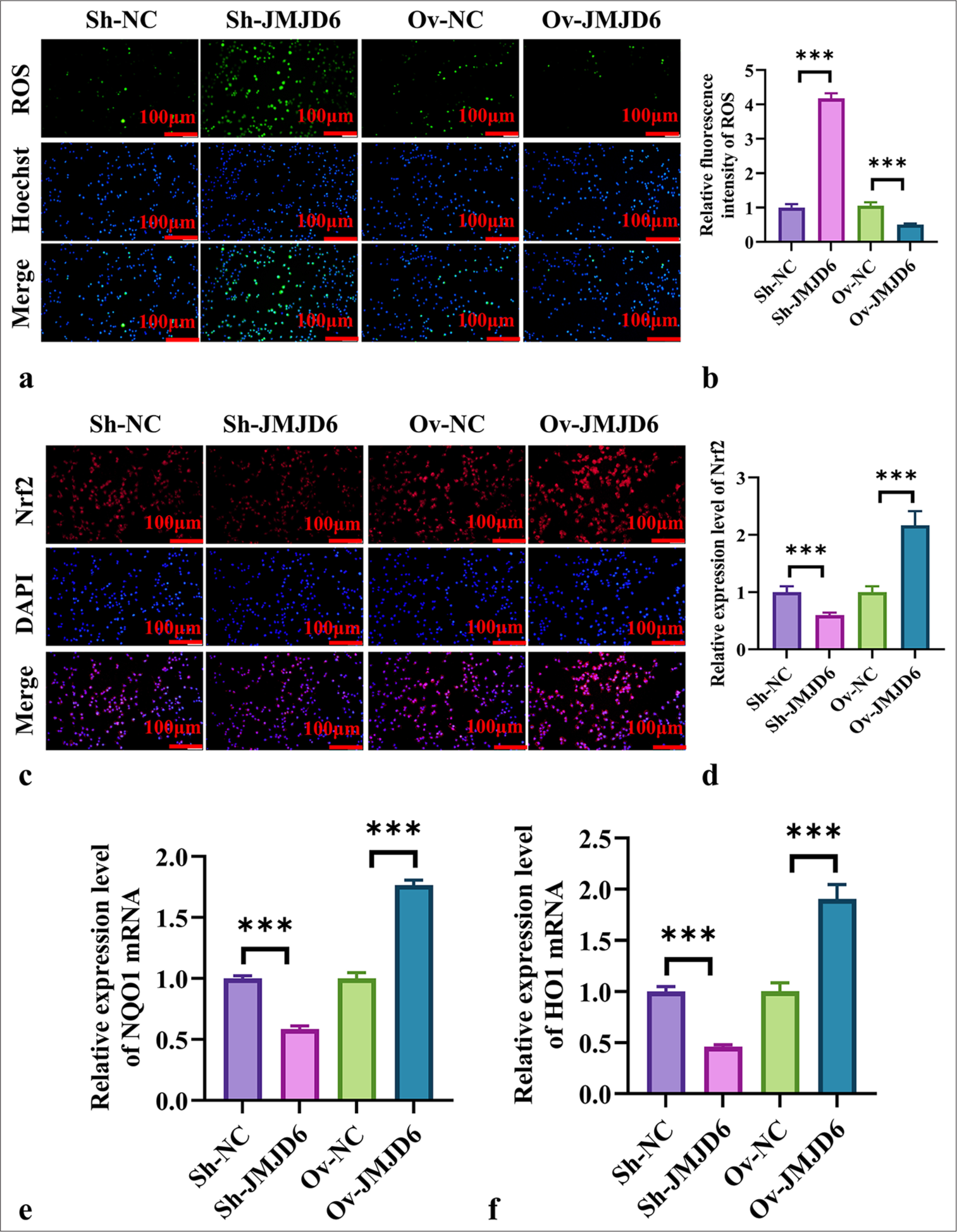
- JMJD6 activates the Nrf2 signaling pathway during autophagy in GC cells. (a and b) Immunofluorescence measurement of ROS levels in MKN-45 cells. (cand d) Immunofluorescence measurement of Nrf2 expression in MKN-45 cells. (e and f) qRT-PCR analysis of NQO1 and HO1 mRNA expression levels. n = 6. ✶✶✶P < 0.001. JMJD6: Jumonji domain–containing protein 6, Sh-NC: ShRNA-negative control, Sh-JMJD6: ShRNA-JMJD6, Ov-NC: Overexpression-negative control, Ov-JMJD6: Overexpression-JMJD6, ROS: Reactive oxygen species, DAPI: 4',6-diamidino-2-phenylindole, Nrf2: Nuclear factor erythroid 2–related factor 2, NQO1: NAD(p)H: quinone oxidoreductase 1, HO1: heme oxygenase-1, qRT-PCR: Quantitative reverse transcription polymerase chain reaction, GC: Gastric cancer, MKN-45: Gastric cancer cell line.
DISCUSSION
In this study, we investigated the expression of JMJD6 in GC and its regulatory effects on PD-L1, EMT, and autophagy. We discovered that JMJD6 promotes the expression of PD-L1 through autophagy activation, thereby enhancing cancer cell growth, migration, infiltration, and EMT. In addition, JMJD6 may activate autophagy by activating the Nrf2 pathway, presenting new insights into the molecular mechanisms of GC. Comparative analysis with other research further strengthens the importance of JMJD6 in cancer progression.
The data from this study suggest that JMJD6 expression is considerably higher in GC cells than in normal cells. This finding is consistent with the results of studies on other cancer types, such as renal cell carcinoma and breast cancers, wherein JMJD6 has been reported to be overexpressed and promote cancer progression.[20,21] Yu et al. demonstrated that Long Intergenic Non-Protein Coding RNA 839 overexpression upregulates JMJD6 mRNA and protein expression, thereby promoting the growth, migration, and invasion of lung cancer cells.[22] Similarly, Zhang et al. found that JMJD6 promotes tumor cell proliferation in osteosarcoma by regulating the cell cycle.[23] In line with these findings, our results show that JMJD6 overexpression notably enhances GC cell growth, migration, and invasion, as demonstrated by CCK-8, EdU, and Transwell assays, whereas JMJD6 knockdown suppresses these oncogenic processes.
We also found that JMJD6 promotes EMT in GC cells by regulating EMT-related markers. JMJD6 overexpression notably increased the levels of the mesenchymal markers N-cadherin, Snail, and Twist while reducing that of the epithelial marker E-cadherin. This finding is consistent with previously reported results, which have shown that in various cancers, JMJD6 promotes metastasis by regulating EMT.[24,25] For example, Liu et al. found that JMJD6 can reverse the effects of CCNB2 downregulation, leading to increased nasopharyngeal carcinoma cell activity in vitro and enhanced tumorigenicity and metastasis in vivo.[24] Our study further confirms that JMJD6 promotes the metastatic potential of GC cells by modulating EMT, indicating that the role of JMJD6 as an EMT regulator is consistent across multiple tumors.
A key finding of this study is the positive correlation between JMJD6 and PD-L1. We found that JMJD6 overexpression considerably increased PD-L1 levels, whereas JMJD6 knockdown reduced PD-L1 expression. PD-L1 is widely recognized as a crucial molecule in cancer immune evasion.[26-28] This study is the first to reveal a novel mechanism by which JMJD6 regulates PD-L1 through autophagy. This finding is a groundbreaking discovery.
Autophagy in cancer plays a dual role: Promoting cell survival and inducing cell death.[29,30] We found that JMJD6 overexpression activated autophagy in GC cells by boosting the levels of LC3II/LC3I and p62, whereas the implementation of autophagy inhibitors, such as 3-MA and Bafilomycin A1, notably reduced PD-L1 expression. This finding suggests that JMJD6 regulates PD-L1 expression through autophagy activation. Moreover, Zhang et al. demonstrated that autophagy regulates PD-L1 expression in lung cancer, promoting the immune evasion of tumor cells.[31] Our study furnishes new evidence for the autophagic regulation of PDL1, further indicating that targeting autophagy to regulate PD-L1 could be a promising therapeutic approach for cancer immunotherapy.
Our study also suggests that JMJD6 may regulate autophagy through the Nrf2 pathway. Nrf2 is a key regulator of oxidative stress, and earlier studies have indicated that Nrf2 activation is closely linked to autophagy activation.[32,33] Consistent with our work, the study of Deng et al. discovered that Nrf2 promotes cancer cell survival in lung cancer by activating autophagy.[34] Similarly, Lu et al. found that in lung cancer, Nrf2 activation protects against oxidative stress-induced cell damage through autophagy.[35] We observed that JMJD6 may promote autophagy by reducing ROS levels through the activation of the Nrf2 pathway. This result further supports the critical role of Nrf2 as a regulator of oxidative stress and autophagy and reveals, for the 1st time, the connection between JMJD6 and Nrf2.
These outcomes contribute new proof for JMJD6 as a possible medical target in GC, particularly with potential applications in combination with autophagy inhibitors or immune checkpoint inhibitors. Future research should further explore the specific molecular mechanisms by which JMJD6 regulates PD-L1 through autophagy and evaluate the therapeutic potential of targeting JMJD6 in preclinical models. In addition, studying the clinical coexpression patterns of JMJD6 and PD-L1 in samples from patients with GC may offer important perspectives on their potential as biomarkers for predicting prognosis and treatment outcomes in GC.
While this study provides valuable insights into the carcinogenic role of JMJD6 in GC, several limitations must be considered. First, the underlying mechanisms through which JMJD6 regulates PD-L1 expression and autophagy activation in GC cells require further validation in clinical samples. The experimental models used in this study, although informative, may not fully replicate the complex tumor microenvironment in vivo. Second, while we observed the involvement of the Nrf2 signaling pathway, additional studies are needed to explore its direct interaction with JMJD6 and its precise role in modulating autophagy in GC. Furthermore, the long-term effects of JMJD6-mediated autophagy activation on tumor progression and treatment resistance remain unclear. Finally, the potential therapeutic implications of targeting JMJD6 in GC require further investigation to assess its effectiveness and safety in preclinical and clinical settings. These limitations highlight the need for future research to elucidate fully the multifaceted role of JMJD6 in GC and its potential as a therapeutic target.
SUMMARY
This study reveals the critical carcinogenic effect of JMJD6 in GC. JMJD6 upregulates PD-L1 expression through autophagy activation, thereby promoting GC cell proliferation, migration, invasion, and EMT. In addition, JMJD6 activates the Nrf2 signaling pathway, which may be related to the regulation of autophagy, further enhancing its carcinogenic potential.
ACKNOWLEDGMENT
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data and materials that support the findings of this study are available from the corresponding author on reasonable request.
ABBREVIATIONS
3-MA: 3-methyladenine
DAPI: 4',6-diamidino-2-phenylindole
E-cadherin: Epithelial cadherin
EdU: 5-ethynyl-2'-deoxyuridine
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
GC: Gastric cancer
GES-1: Gastric epithelial cell line
HO1: Heme oxygenase 1
JMJD6: Jumonji domain–containing protein 6
LC3: Microtubule-associated protein 1A/1B-light chain 3
MKN-45: Gastric cancer cell line
N-cadherin: Neural cadherin
NQO1: NAD(P)H quinone dehydrogenase 1
Nrf2: Nuclear factor erythroid 2-related factor 2
Ov-JMJD6: Overexpression-JMJD6
Ov-NC: Overexpression-negative control
p62: Sequestosome 1
PD-L1: Programmed death-ligand 1
ROS: Reactive oxygen species
Sh-JMJD6: ShRNA-JMJD6
Sh-NC: ShRNA-negative control
Snail: Snail family transcriptional repressor 1
Twist: Twist family BHLH transcription factor 1
AUTHOR CONTRIBUTIONS
XYZ: Conducted the research and contributed to data analysis and interpretation of the results; DN: Provided assistance and suggestions for the experiments. All authors participated in the drafting and critical revision of the manuscript. All authors have read and approved the final manuscript. All authors were fully involved in the work, able to take public responsibility for relevant portions of the content, and agreed to be accountable for all aspects of the work, ensuring that any questions related to its accuracy or integrity are addressed.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study does not involve human or animal research, so there is no need to provide animal ethics or clinical ethics approval. Since this study does not involve human subjects, informed consent is not required.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- Gastric intestinal metaplasia: Real culprit or innocent bystander as a precancerous condition for gastric cancer? Gastroenterology. 2023;165:1352-66.e1.
- [CrossRef] [PubMed] [Google Scholar]
- Predictive biomarkers for immunotherapy in gastric cancer: Current status and emerging prospects. Int J Mol Sci. 2023;24:15321.
- [CrossRef] [PubMed] [Google Scholar]
- Tumor recurrence and survival prognosis in patients with advanced gastric cancer after radical resection with radiotherapy and chemotherapy. World J Gastrointest Surg. 2024;16:1660-9.
- [CrossRef] [PubMed] [Google Scholar]
- Oncogenic DDX46 promotes pancreatic cancer development and gemcitabine resistance by facilitating the JMJD6/CDK4 signaling pathway. Neoplasma. 2024;71:231-42.
- [CrossRef] [PubMed] [Google Scholar]
- Metabolic reprogramming of cancer cells by JMJD6-mediated pre-mRNA splicing associated with therapeutic response to splicing inhibitor. Elife. 2024;12:RP90993.
- [CrossRef] [PubMed] [Google Scholar]
- Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6. Proc Natl Acad Sci U S A. 2022;119:e2201483119.
- [CrossRef] [PubMed] [Google Scholar]
- The JMJD6/HURP axis promotes cell migration via NF-κB-dependent centrosome repositioning and Cdc42-mediated Golgi repositioning. J Cell Physiol. 2022;237:4517-30.
- [CrossRef] [PubMed] [Google Scholar]
- PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;22:1264-75.
- [CrossRef] [PubMed] [Google Scholar]
- The ubiquitin E3 ligase FBXO22 degrades PD-L1 and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci U S A. 2021;118:e2112674118.
- [CrossRef] [PubMed] [Google Scholar]
- Regulation of PD-L1 expression in the tumor microenvironment. J Hematol Oncol. 2021;14:10.
- [CrossRef] [PubMed] [Google Scholar]
- EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol Cancer. 2019;18:165.
- [CrossRef] [PubMed] [Google Scholar]
- METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21:60.
- [CrossRef] [PubMed] [Google Scholar]
- ILT4 inhibition prevents TAM-and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation. Theranostics. 2021;11:3392-416.
- [CrossRef] [PubMed] [Google Scholar]
- LSD1 deletion decreases exosomal PD-L1 and restores T-cell response in gastric cancer. Mol Cancer. 2022;21:75.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy: Regulator of cell death. Cell Death Dis. 2023;14:648.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. 2023;24:560-75.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy and mitophagy in cancer metabolic remodelling. Semin Cell Dev Biol. 2020;98:129-38.
- [CrossRef] [PubMed] [Google Scholar]
- Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis. 2022;1868:166400.
- [CrossRef] [PubMed] [Google Scholar]
- Both EZH2 and JMJD6 regulate cell cycle genes in breast cancer. BMC Cancer. 2020;20:1159.
- [CrossRef] [PubMed] [Google Scholar]
- An oncogenic JMJD6-DGAT1 axis tunes the epigenetic regulation of lipid droplet formation in clear cell renal cell carcinoma. Mol Cell. 2022;82:3030-44.e8.
- [CrossRef] [PubMed] [Google Scholar]
- LINC00839/miR-519d-3p/JMJD6 axis modulated cell viability, apoptosis, migration and invasiveness of lung cancer cells. Folia Histochem Cytobiol. 2021;59:271-81.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-298 suppresses the malignant progression of osteosarcoma by targeting JMJD6. Eur Rev Med Pharmacol Sci. 2022;26:2250-8.
- [Google Scholar]
- Cyclin B2 impairs the p53 signaling in nasopharyngeal carcinoma. BMC Cancer. 2024;24:25.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-770 inhibits tumorigenesis and EMT by targeting JMJD6 and regulating WNT/β-catenin pathway in non-small cell lung cancer. Life Sci. 2017;188:163-71.
- [CrossRef] [PubMed] [Google Scholar]
- SMARCAL1 is a dual regulator of innate immune signaling and PD-L1 expression that promotes tumor immune evasion. Cell. 2024;187:861-81.e32.
- [CrossRef] [PubMed] [Google Scholar]
- Acidosis-mediated increase in IFN-γ-induced PD-L1 expression on cancer cells as an immune escape mechanism in solid tumors. Mol Cancer. 2023;22:207.
- [CrossRef] [PubMed] [Google Scholar]
- Hsa_ circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC Axis and PD-L1 phosphorylation. Mol Cancer. 2023;22:205.
- [CrossRef] [PubMed] [Google Scholar]
- HMGB1 in the interplay between autophagy and apoptosis in cancer. Cancer Lett. 2024;581:216494.
- [CrossRef] [PubMed] [Google Scholar]
- Selective autophagy in cancer: Mechanisms, therapeutic implications, and future perspectives. Mol Cancer. 2024;23:22.
- [CrossRef] [PubMed] [Google Scholar]
- CDK5 destabilizes PD-L1 via chaperon-mediated autophagy to control cancer immune surveillance in hepatocellular carcinoma. J Immunother Cancer. 2023;11:e007529.
- [CrossRef] [PubMed] [Google Scholar]
- FGF2 Rescued cisplatin-injured granulosa cells through the NRF2-autophagy pathway. Int J Mol Sci. 2023;24:14215.
- [CrossRef] [PubMed] [Google Scholar]
- Co-targeting autophagy and NRF2 signaling triggers mitochondrial superoxide to sensitize oral cancer stem cells for cisplatin-induced apoptosis. Free Radic Biol Med. 2023;207:72-88.
- [CrossRef] [PubMed] [Google Scholar]
- PI3K/mTOR inhibitors promote G6PD autophagic degradation and exacerbate oxidative stress damage to radiosensitize small cell lung cancer. Cell Death Dis. 2023;14:652.
- [CrossRef] [PubMed] [Google Scholar]
- Hsa_circ_0096157 silencing suppresses autophagy and reduces cisplatin resistance in non-small cell lung cancer by weakening the Nrf2/ARE signaling pathway. Mol Biol Rep. 2024;51:703.
- [CrossRef] [PubMed] [Google Scholar]








