Translate this page into:
Upregulation of phosphatase and tensin homolog deleted on chromosome ten inhibits lung cancer cell proliferation by suppressing the oncogene polo-like kinase 1 and inducing autophagy

*Corresponding author: Limin Huang, Department of Oncology, Weifang Traditional Chinese Medicine Hospital, Weifang, Shandong, China. hlmhappy@163.com
-
Received: ,
Accepted: ,
Abstract
Objective:
Lung cancer is one of the main causes of cancer-related mortality globally, and it poses considerable therapeutic challenges. Polo-like kinase 1 (PLK1) exhibits upregulation in lung cancer, and PLK1 silencing promotes autophagy in lung cancer cells, which inhibits tumor progression. The phosphatase and tensin homolog deleted on chromosome ten (PTEN) acts as a tumor suppressor gene. This study aimed to investigate whether PTEN regulates autophagy and inhibits lung cancer-cell proliferation by suppressing PLK1.
Material and Methods:
In this study, we evaluated cell proliferation by silencing or overexpressing PLK1 and PTEN in A549 cells through 5-ethynyl-2'-deoxyuridine labeling and cloning experiments. The autophagy levels were detected through transmission electron microscopy, real-time quantitative polymerase chain reaction, and Western blot. Finally, the results of in vitro experiment were further verified using an in vivo xenograft tumor animal model.
Results:
The upregulation of PTEN suppressed PLK1 expression in lung cancer cells and reduced their proliferation rate. In addition, the overexpression of PTEN has been associated with the growth of lung cancer tumors. In parallel, the levels of autophagy of lung cancer cells rose in response to PTEN upregulation in vivo and in vitro.
Conclusion:
This study revealed that PTEN promotes the autophagy of lung cancer cells and inhibits cell proliferation and tumor growth by suppressing PLK1 expression. This finding provides a new strategy for lung cancer treatment by utilizing the autophagy-regulating effect of PTEN to inhibit lung cancer growth by targeting PLK1.
Keywords
Autophagy
Cell proliferation
Lung cancer
Phosphatase and tensin homolog deleted on chromosome ten
Polo-like kinase 1
INTRODUCTION
Lung cancer has been a focus of medical research for its challenging treatment.[1,2] Despite advances in lung cancer therapy, such as targeted and immune therapies, the 5-year survival rate remains low.[3,4] Thus, new therapeutic approaches and strategies must be explored.[5,6] The phosphatase and tensin homolog deleted on chromosome ten (PTEN) gene in lung cancer acts as a tumor suppressor gene.[7] The phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway is excessively activated in lung cancer due to the mutations, deletions, or downregulation of the PTEN gene. This pathway promotes the growth, survival, and metastatic potential of tumor cells, which speeds up disease progression and deterioration.[8] The PTEN gene also performs a key function in the prevention of the growth of tumor cells by controlling apoptosis and the course of the cell cycle.[9] Therefore, comprehending and intervening in the lung cancer mechanisms of PTEN is important for the development of new therapeutic strategies and prognosis assessment.
With a greater understanding of tumor biology, polo-like kinase 1 (PLK1), a critical protein in cell cycle regulation, has become essential to tumor growth.[10,11] Abnormally elevated expression levels of PLK1 have been closely associated with the invasiveness, malignancy, and prognosis of various cancers.[12] PLK1 inhibitors can disrupt the rapid proliferation of non-small cell lung cancer cells and trigger them to undergo apoptosis.[13-15] As a route for intracellular breakdown, autophagy contributes to the preservation of cellular homeostasis and averting external stress.[16] Autophagy plays a dual role in tumorigenesis and development, which both limit the initial stages of tumors and potentially promote tumor cell adaptation to environmental stress.[17,18] The activation of the autophagy pathway inhibits lung cancer cell proliferation, which suggests that autophagy may be a potential target for lung cancer therapy.[19] However, no research reported the effect of PTEN on PLK1 expression and its anti-lung cancer effects through the regulation of autophagy.
Although the antitumor effects of PTEN have been widely studied, its specific molecular mechanisms in lung cancer remain unclear.[20] Particularly, whether PTEN can exert its antitumor effects by inhibiting PLK1 and whether such a process involves autophagy regulation have not been explored. Furthermore, the specific role of autophagy in lung cancer cells, PTEN’s mechanism in the precise regulation of autophagy, and how these molecular events collectively inhibit lung cancer cell proliferation are pressing issues that need to be addressed. Therefore, the exploration of the mechanisms of PTEN in lung cancer treatment, especially its effects on PLK1 and autophagy, is important for the development of new lung cancer treatment strategies.
Based on the above background, this study aimed to investigate the mechanisms of PTEN in lung cancer treatment, particularly its effect on PLK1 expression and whether it suppresses lung cancer cell proliferation through autophagy pathway regulation. Through in vitro cell experiments and molecular biology techniques, this study elucidated the effects of PTEN on lung cancer cell proliferation, explored the regulatory role of PTEN in PLK1 expression, and revealed the potential mechanisms of PTEN in autophagy activation. The findings of this study will provide a theoretical basis for novel lung cancer treatment strategies, enrich the understanding of PTEN’s antitumor mechanisms, and possibly uncover new targets for lung cancer therapy.
MATERIAL AND METHODS
Cell culture and grouping treatment
BEAS-2B cells (FN6080086) and A549 cells (BFN60800665) were obtained from the American Type Culture Collection (Manassas, VA, USA). Mycoplasma detection and short tandem repeat identification were carried out, and non-pollution results were obtained. The cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 (12633020, Gibco, Life Technologies, Rockville, MD, USA) with 10% fetal bovine serum (C0235, GIBCO BRL, Grand Island, NY, USA) and 1% antibiotics. Afterward, cell morphology was observed under an inverted microscope (Ti2-E, Nikon, Tokyo, Japan). The cell grouping and treatment methods were as follows: negative control to PLK1 siRNA (si-NC), PLK1 siRNA (siPLK1), control, negative control to overexpression plasmid (Ov-NC), PTEN overexpression plasmid (Ov-PTEN), PLK1 overexpression plasmid (Ov-PLK1), PLK1 overexpression plasmid+ negative control to overexpression plasmid (OvPLK1+Ov-NC), and PTEN and PLK1 overexpression plasmid (Ov-PLK1+ Ov-PTEN).
Cell transfection
Before siRNA or plasmid transfection, the tumor cells were cultured in culture dishes containing an appropriate growth medium. The cells should be in the logarithmic growth phase. siRNA PLK1 and transfection reagent (Lipofectamine 2000, 11668019, Thermo Fisher Scientific, Waltham, MA, USA) were mixed separately with serum-free culture medium and incubated at room temperature for a certain period to form the siRNA–Lipofectamine mixture. The siRNA– Lipofectamine mixture was slowly added to the tumor cell-culture dish containing serum-free culture medium. The dish was gently shaken to ensure the even distribution of the mixture on the cells. Transfection was conducted for 6 h. After transfection, the transfection mixture was replaced with a serum-containing culture medium. Experiments were performed 24–72 h after transfection. The effectiveness of siRNA transfection was evaluated through assessment of the target gene through Western blot. The transfection plasmid sequence is as follows:
PLK1 siRNA: 5'-UAUUCAUUCUUCUUGAUCCGG-3'
PTEN overexpression plasmid: Forward: 5'-CCGGAATTCA TGGCCATGGCAACCAAAGG-3'; and reverse: 5'-CCCAA GCTTTCAGACTTTTGTAATTTGTGTATGC-3'.
PLK1 overexpression plasmid: CCACCAGCACGTCGTA GGATTCCACGGCTTTTTCGAGGACAACGACTTCG TGTTCGTGGTGTTGGAGCTCTGCCGCCGGAGGT CTCTCCTGGAGCTGCACAAGAGGAGGAAAGCCCT GACTGAGCCTGAGGCCCGATACTACCTACGGCAAAT TGTGCTTGGCTGCCAGTACCTGCACCGAAACCGA GTTATTCATCGAGACCTCAAGCTGGGCAACCTT TTCCTGAATGAAGATCTGGAGGTGAAAATAGGGGAT TTTGGACTGGCAACCAAAGTCGAATATGACGGG GAGAGGAAGAAGACCCTGTGTGGGACTCCTAATTA CATAGCTCCCGAGGTGCTGAGCAAGAAAGGGCA CAGTTTCGAGGTGGATGTGTGGTCCATTGGGTG TATCATGTATACCTTGTTAGTGGGCAAACCAC CTTTTGAGACTTCTTGCCTAAAAGAGACCTACCTC CGGATCAAGAAGA.
Bioinformatic analysis
Gene expression profiling interactive analysis (GEPIA) is an online tool used for the analysis of gene expression and patient survival data. The procedures for utilizing the GEPIA website to analyze the PLK1 gene and its association with a lung cancer patient’s prognosis are listed below. Open the GEPIA website (http://gepia.cancer-pku.cn/). Enter “PLK1” in the search box and select “PLK1” gene for analysis. In the “Survival Map” tab, select “Overall Survival” to examine the relationship between PLK1 gene expression and the overall survival or disease-free survival of lung cancer patients. Analyze the connection between a lung cancer patient’s prognosis and PLK1 gene expression.
Reactive oxygen species (ROS) detection
A549 cells (1 × 106 cells/well) were seeded in 24-well plates (cat no. 07-6024, MACGENE, Beijing, China). Cells were treated for 24 h and then incubated with 5 mM H2DCFDA (D399, Thermo Fisher Scientific, Waltham, MA, USA). The nucleus was stained by 4’,6-Diamidino-2’-phenylindole (DAPI, C0065, Solarbio, Beijing, China). Fluorescence microscopy was performed to detect the intensity of the DCF fluorescence.
Western blot assay
The collected cells were mixed well with radioimmunoprecipitation assay (RIPA, P0013J, Beyotime Biotechnology, Shanghai, China). After centrifugation at 4°C, the supernatant was aspirated, and a bicinchoninic acid assay (BCA) kit was used to measure the amount of total protein present (P0010S, Beyotime Biotechnology, Shanghai, China). All protein samples were separated and attached to the membranes. Then, the membranes were co-incubated at 4°C with primary antibodies, including the following: Nuclear factor erythroid 2-related factor 2 (1:1000 dilution, 80593-1-RR, Proteintech, Wuhan, Hubei, Chian), light chain 3 (LC3)-I (1:5000 dilution, 18722-1-AP, Proteintech), LC 3-II (1:1000 dilution, 18725-1-AP, Proteintech), Beclin 1 (1:1000 dilution, 11306-1-AP, Proteintech, Wuhan, Hubei, Chian), autophagy protein 5 (ATG5) (1:1000 dilution, 10181-2-AP, Proteintech, Wuhan, Hubei, China), and glyceraldehyde-3-phosphate dehydrogenase (1:1000 dilution, TA-08, ZSGBbio, Beijing, China). Secondary antibody horseradishperoxidase (HRP) goat antimouse immunoglobulin G (IgG) (1:2000 dilution, ZB-2305, ZSGB-bio, Beijing, China) or HRP goat antirabbit IgG (1:2000 dilution, ZB-2301, ZSGB-bio, Beijing, China) was added and incubated to the membranes at room temperature. Finally, the bands were measured using an enhanced chemiluminescence kit (BL520b, Biosharp Life Science, Hefei, Anhui, China) and Image Quant LAS4000 (GE Healthcare, Chicago, IL, USA), followed by analysis using ImageJ software (Fiji software version 2.0, LOCI, University of Wisconsin, Madison, Wisconsin, USA).
5-Ethynyl-2'-deoxyuridine (EdU) assay
A total of 20 µM EdU working solution (C0071S, Beyotime, Shanghai, China) was added to the culture of an appropriate number of cells. Then, the cells were fixed with 4% paraformaldehyde (P0099, Beyotime, Shanghai, China). Next, 300 µL 0.5% Triton X-100 (T8200, Solarbio, Beijing, China) was added to each well. Each well was added with 0.5 mL click reaction solution. DAPI (C0060, Beyotime, Shanghai, China) staining solution was added dropwise for incubation in the dark for 10 min. Then, the wells were shaken with phosphate-buffered saline (PBS) and washed thrice. An antifluorescence quenching agent (S2100, Solarbio, Beijing, China) was added to seal the tablets. The collected images were observed under a fluorescence microscope (CKX53, OLYMPUS, Tokyo, Japan). The percentage of EdU-positive cells was assessed using ImageJ software (1.48, National Institutes of Health, Rockville, Maryland, USA).
Colony formation assay
Cells were digested with trypsin, centrifuged to obtain a cell suspension, and continuously cultured for 10 days at a concentration of 1000/well. Next, the cells were fixed with 4% paraformaldehyde for 30 min. Afterward, 1 mL of 0.1% crystal violet dye solution was added to each hole. Dyeing was conducted for 10–30 min, and the dye solution was discarded. The cells were washed and dried, and their photos were captured and recorded. The colonies were counted through ImageJ software (1.48, National Institutes of Health, Rockville, Maryland, USA).
Transmission electron microscopy
An electron microscope fixative (G1102, Servicebio, Wuhan, China) was added to the cell precipitate, which was resuspended at 4°C for 2–4 h. After the addition of 0.1 M phosphate buffer (pH 7.4), the cells were rinsed and centrifuged. Then, 1% agarose solution was added to the precipitate to wrap it up. After being fixed for 2 h at room temperature in the dark with 1% osmic acid (Ted Pella, Inc., California, USA), the samples were rinsed thrice with PBS. Using gradient concentration of alcohol dehydration, 100% acetone treatment was conducted twice. At 37°C, the mixture of acetone and 812 embedding agent (90529-77-4, SPI) was used in overnight penetration. Subsequently, the sample was inserted into the embedding plate and treated overnight with a pure 812 embedding agent. ImageJ software (1.48, National Institutes of Health, Rockville, Maryland, USA) was used in the examination of images under a transmission electron microscope (HT7800/HT7700, HITACHI, Japan).
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using an RNA simple kit (DP419), and the RNA was reverse-transcribed into complementary DNA using the Quant complementary DNA (cDNA) first-strand synthesis kit (KR103). Fast Fire reagent (SYBR Green, FP207) was used during a quantitative polymerase chain reaction to detect the messenger RNA (mRNA) expressions of genes displayed in Table 1 using the above primers. β-Actin gene expression was used as the endogenous control, and the (2-∆∆Ct) ratio was used to calculate the expression level. All the above reagents were purchased from Tiangen (Beijing, China).
| Primer | Primer sequences-forward (5’-3’) | Primer sequences-reverse (5’-3’) |
|---|---|---|
| Human | ||
| Beclin | GGCTGAGAGACTGGATCAGG | CTGCGTCTGGGCATAACG |
| P62 | ATCGGAGGATCCGAGTGT | TGGCTGTGAGCTGCTCTT |
| ATG5 | AAAGATGTGCTTCGAGATGTGT | CACTTTGTCAGTTACCAACGTCA |
| PLK1 | CAGCAAGTGGGTGGACTATT | GAGGATGAGGCGTGTTGAGT |
| PTEN | TGGGCCCTGTACCATCCCAAGT | TGTGGCAACCACAGCCATCGT |
| β-actin | TCTGGCAACGGTGAAGGTGACA | CACCTCCCCTGTGTGGACTT |
| Mouse | ||
| Beclin | GAGTCTAGACTCGTGGTGGA | GAGTCTAGACTCGTGGTGGA |
| P62 | GCTCTTCGGAAGTCAGCAAACC | GCAGTTTCCCGACTCCATCTGT |
| ATG5 | TGTGCTTCGAGATGTGTGGTT | GTCAAATAGCTGACTCTTGGCAA |
| β-actin | TGTTACCAACTGGGACGACA | GGGGTGTTGAAGGTCTCAAA |
ATG5: Autophagy protein 5, PLK1: Polo-like kinase 1, PTEN: Phosphatase and tensin homolog deleted on chromosome ten, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Animals
A total of 24 male Blab/c nude mice (4–6 weeks) were provided by Beijing Vital River Laboratory Animal Technology Co., Ltd (SCXK [Jing] 2016–0006). The animals were kept in a specific pathogen-free environment where they ate and drank freely. After 1 week of acclimation, the mice were randomly divided into four groups: Negative control A549 cell (NC), overexpression of PLK1 A549 cell (PLK1), overexpression of PLK1 + negative control A549 cell (PLK1 + NC), and overexpression of PLK1 + overexpression of PTEN A549 cell (PLK1 + PTEN) groups. Each group of mice was injected with 2 × 106 cells in their right axillary region. The tumor’s long and short diameters were measured, and its growth was routinely monitored. The mice were killed through cervical dislocation 4 weeks later. The tumors were collected, weighed, and captured on camera. Tumor volume was calculated as long diameter × short diameter2/2. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Hospital. The study received approval from the Institutional Animal Care and Use Committee of Hospital.
Hematoxylin and eosin (H&E) staining
The collected tumor tissues were fixed with 4% paraformaldehyde and were sectioned (4 µm) after paraffin embedding. The images were stained using a H&E Stain Kit (G1120, Solarbio, Beijing, China) and observed under a microscope (CX41-32RFL, Olympus, Tokyo, Japan).
Statistical analysis
Statistical analysis was conducted with the Statistical Package for the Social Sciences 20.0 software (Analysis Software [Shanghai] Co., Ltd., Shanghai, China). All data were presented as mean ± standard deviation from three independent experiments. Multiple group comparisons were conducted using analysis of variance, followed by Bonferroni’s post hoc test, with α = 0.05. Comparisons between the two groups were analyzed through an independent sample t-test. Data analysis was performed using GraphPad 8.0.2 software (San Diego, California, USA https://www.graphpad-prism.cn/). Statistical significance was considered at P < 0.05.
RESULTS
Upregulation of PLK1 in Lung Cancer and Silencing of PLK1 Inhibits Cell Proliferation
The analysis of PLK1 expression in lung cancer tissues revealed PLK1 in lung cancer tissues [Figure 1a]. High PLK1 expression may be linked to a poor prognosis of individuals with lung cancer, according to a correlation study between PLK1 expression and prognosis. This finding provides important clues for further research on PLK1 as a potential biomarker for lung cancer prognosis and lays the foundation for the clinical application of PLK1 as a therapeutic target [Figure 1b]. The efficiency verification results of si-PLK1 transfection show the lower PLK1 levels in the si-PLK1 group. This finding indicates that si-PLK1 transfection successfully inhibited the expression of PLK1 (P < 0.01) [Figure 1c-e] and thus lays the foundation for further research on the function and mechanism of action of PLK1 in lung cancer. The experimental results in Figure 1f-i reveal that silencing of PLK1 inhibited the proliferation of lung cancer cells, with the proliferation ability of the si-PLK1 group being lower (P < 0.01) (P < 0.001). This outcome implies that PLK1 silencing may have an inhibitory effect on lung cancer cell growth and multiplication.
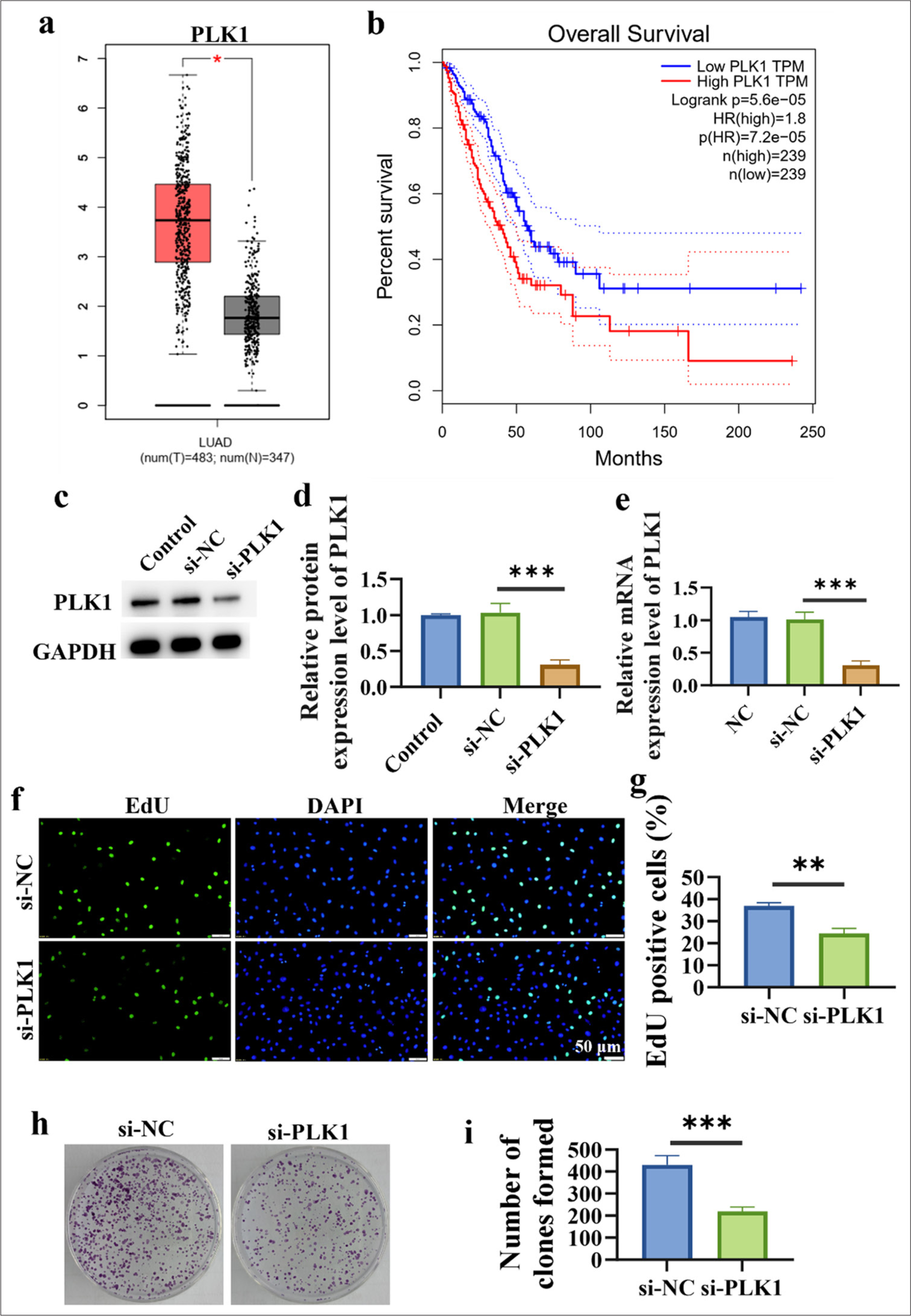
- Upregulation of PLK1 in Lung Cancer and Inhibition of Cell Proliferation by Silencing PLK1. (a) Analysis of PLK1 expression levels in lung cancer. (b) Correlation analysis between PLK1 expression and prognosis of lung cancer patients. (c-e) Validation of si-PLK1 transfection efficiency. (f and g) Inhibition of lung cancer cell proliferation by silencing PLK1. (h and i) Inhibition of clonogenic capacity in lung cancer cells by silencing PLK1. n = 3. (*P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001). PLK1: Polo-like kinase 1, si-NC: negative control to PLK1 siRNA, si-PLK1: PLK1 siRNA.
Silencing of PLK1 promotes autophagy in lung cancer cells and inhibits lung cancer progression
In cells, the production and clearance of ROS are in a dynamic balance, with moderate levels of ROS promoting the maintenance of normal cellular physiological functions, whereas excessive levels of ROS may lead to cell damage and apoptosis. Figures 2a and b show the higher ROS content in the si-PLK1 group, which indicates that silencing of PLK1 promotes the increase in ROS levels within lung cancer cells (P < 0.01). This finding also implies that PLK1 silencing may affect the biological behavior of lung cancer cells by controlling ROS levels, which in turn may have influenced the onset and spread of lung cancer. The experimental findings in Figures 2c and d reveal the higher levels of LC3 II/I (P < 0.05), Beclin 1 (P < 0.001), and ATG5 (P < 0.01) in the si-PLK1 group. Meanwhile, the level of P62 (P < 0.05) was lower than that in the si-NC group. These findings suggest that in lung cancer cells, PLK1 knockdown enhances the expression of proteins linked to autophagy. Autophagosomes refer to the vesicular structures formed during the autophagy process, and they contain degraded organelles and proteins. The formation and accumulation of autophagosomes serve as important indicators of the autophagy process, which reflect the intensity of autophagic activity within cells. Electron microscopy experimental finding unveils the higher number of autophagosomes in the si-PLK1 group, which implies that silencing of PLK1 promotes the accumulation of autophagosomes in lung cancer cells [Figure 2e].
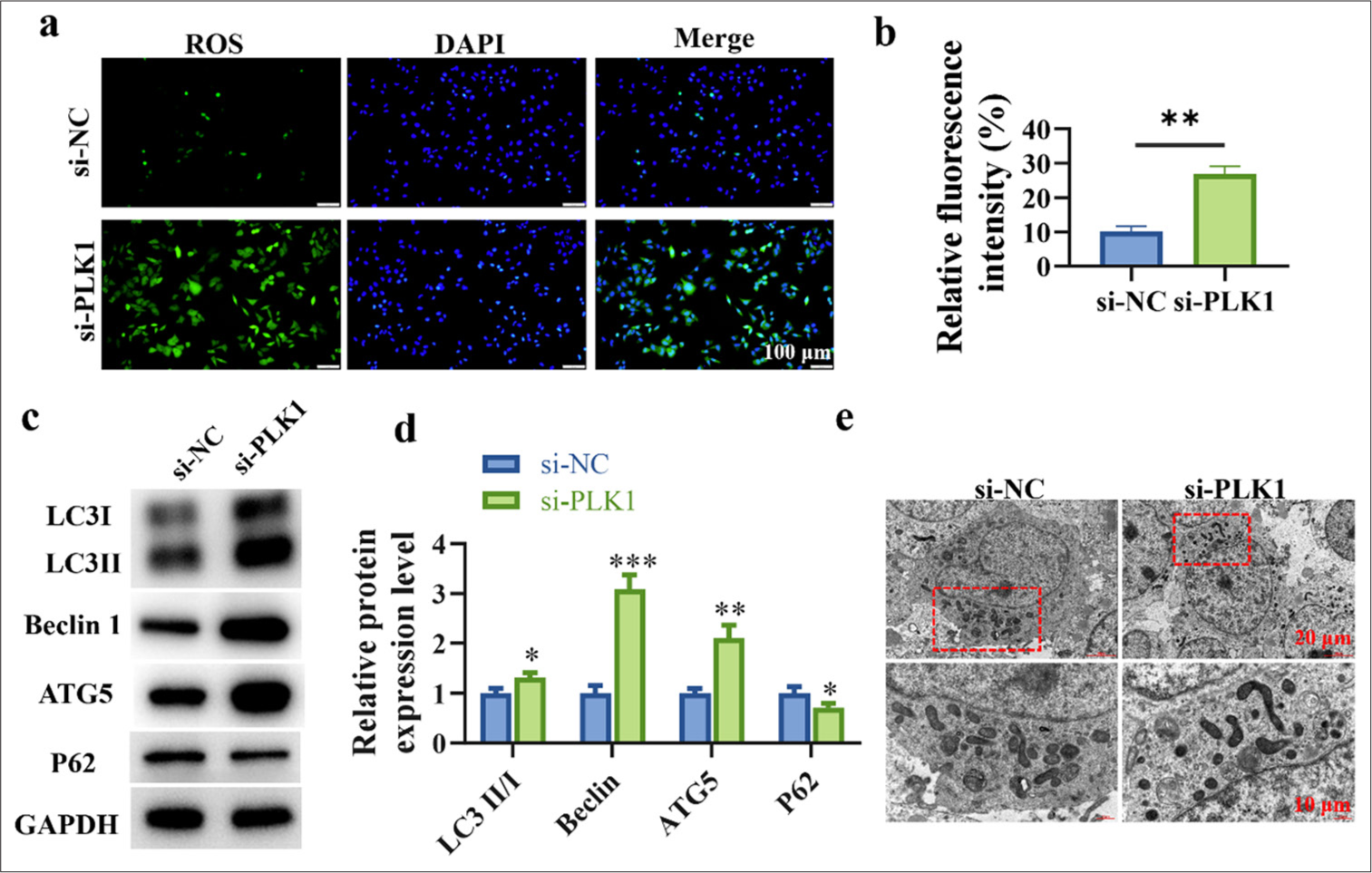
- Silencing of PLK1 promotes autophagy in lung cancer cells and inhibits cancer progression. (a and b) Induction of ROS increase in lung cancer cells through silencing of PLK1. (c and d) Induction of autophagy in lung cancer cells through silencing of PLK1. (e) Accumulation of autophagosomes in lung cancer cells through PLK1 silencing. n = 3 (✶P < 0.05, ✶✶P < 0.01, ✶✶✶ P < 0.001). PTEN: Phosphatase and tensin homolog deleted on chromosome ten, ATG5: Autophagy protein 5, PLK1: Polo-like kinase 1, ROS: Reactive oxygen species, Ov-NC: negative control to overexpression plasmid, Ov-PTEN: PTEN overexpression plasmid.
PTEN expression is downregulated in lung cancer cells
Figure 3a-c shows that A549 cells displayed a substantial decrease in PTEN mRNA and protein expressions compared with BEAS-2B cells (P < 0.05). Subsequently, PTEN was overexpressed, and the overexpression efficiency was verified through Western blot and RT-qPCR. Figure 3d-f reveals the more efficient overexpression of PTEN (P < 0.001).
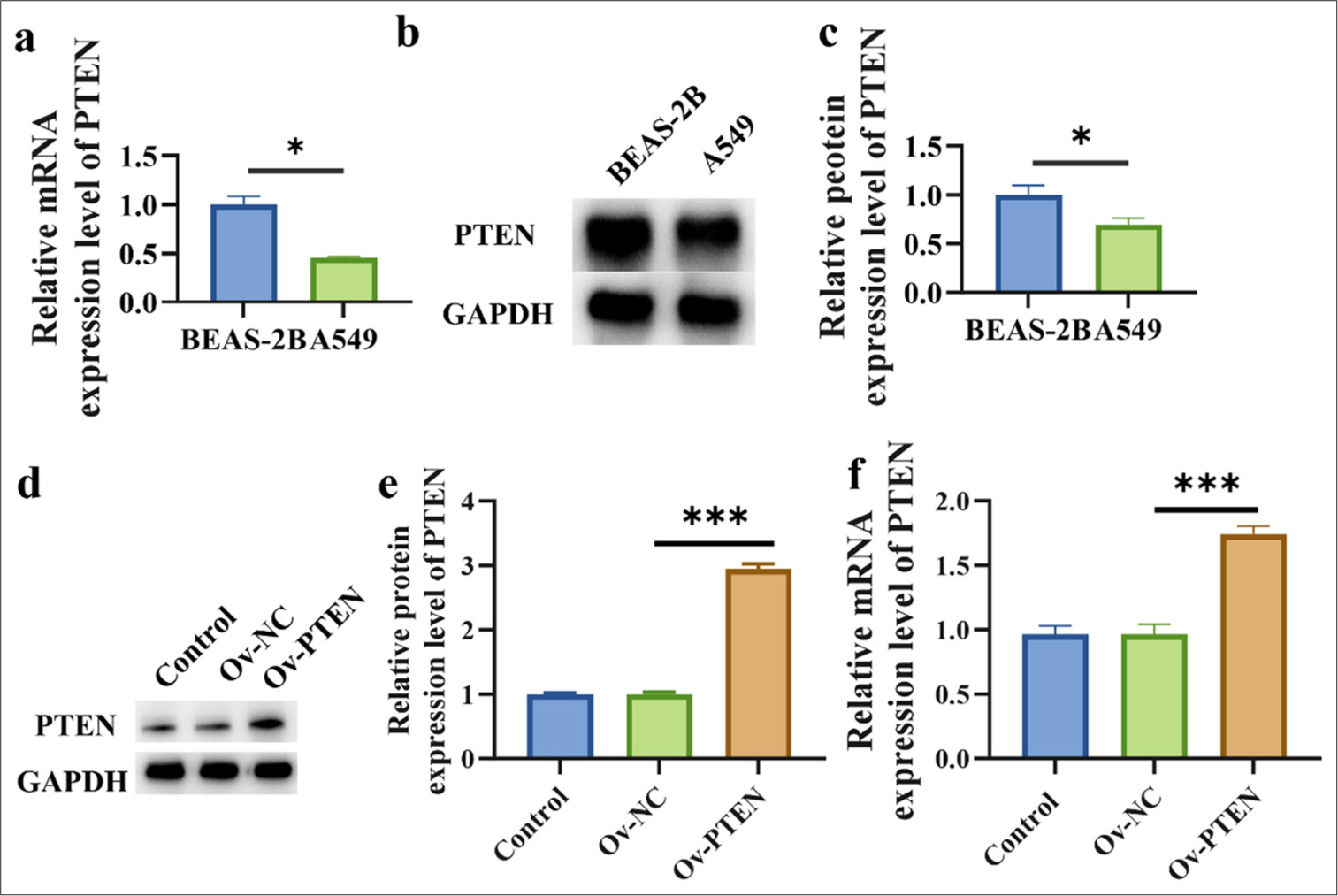
- PTEN expression is reduced in lung cancer cells. (a) Transcription level of PTEN. (b and c) Protein level of PTEN. (d and e) PTEN protein expression level after PTEN overexpression in A549 cells. (f) PTEN mRNA expression level after PTEN overexpression in A549 cells. n=3. (✶P < 0.05, ✶✶✶P < 0.001).
Overexpression of PTEN promotes autophagy in lung cancer cells and inhibits lung cancer cell proliferation
The experimental results in Figures 4a and b indicate that PTEN overexpression significantly increased intracellular ROS levels, which suggests that PTEN upregulation promoted ROS production within cells (P < 0.001). LC3 II/I (P < 0.05), Beclin 1 (P < 0.001), and ATG5 (P < 0.05) are proteins closely related to the autophagy process, and their increased expressions suggest that PTEN upregulation enhanced the expression of autophagy-related proteins in lung cancer cells. The decreased expression of P62 (P < 0.05) supports this conclusion, which indicates that PTEN upregulation promotes autophagy in lung cancer cells [Figure 4c and d]. Electron microscopy results unveil that PTEN upregulation promoted autophagosome accumulation in lung cancer cells, which further supports the notion that PTEN upregulation enhances autophagy in lung cancer cells [Figure 4e]. Cell proliferation is an important biological characteristic of lung cancer cells. The findings indicate that after PTEN upregulation, the proliferation and colony-forming ability of lung cancer cells decreased, which means that PTEN upregulation can inhibit the proliferation of lung cancer cells (P < 0.001) [Figure 4f-i].
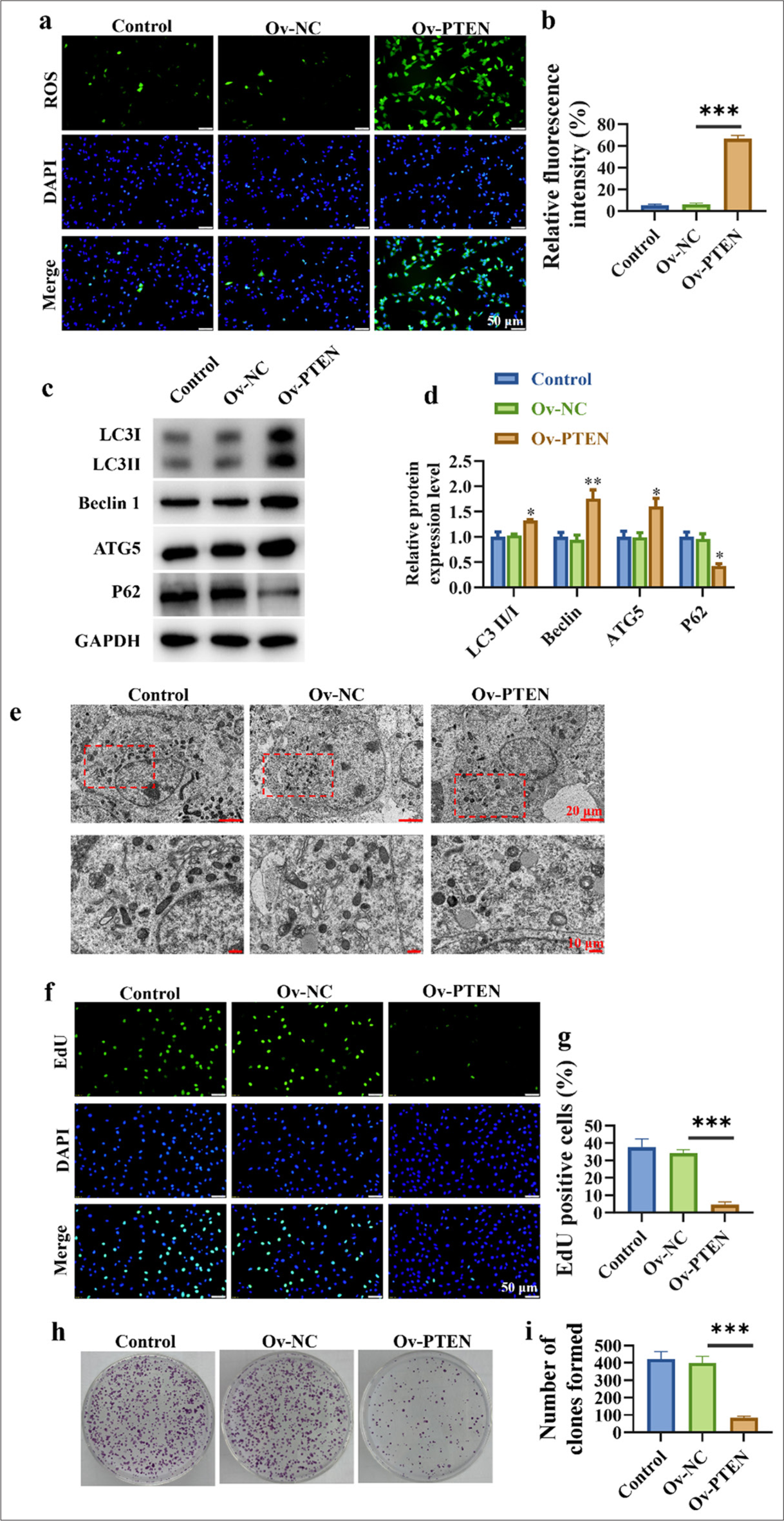
- Up-regulation of PTEN promotes autophagy and inhibits the proliferation of lung cancer cells. (a and b) Detection of ROS levels in cells. (c and d) Promotion of autophagy-related protein expression in lung cancer cells. (e) Accumulation of autophagosomes in lung cancer cells. (f and g) Dose-dependent inhibition of lung cancer cell proliferation. (h and i) Dose-dependent inhibition of clonogenic capacity in lung cancer cells. n=3 (✶P < 0.05, ✶✶ P < 0.01, ✶✶✶ P < 0.001).
Up-regulation of PTEN inhibits PLK1
PLK1 is an oncogene typically overexpressed in tumor cells. Figures 5a and b show the decreased expression level of PLK1 in the Ov-PTEN group (P < 0.001). These findings suggest that PTEN upregulation can exert its anticancer effects by inhibiting the expression of PLK1. Subsequently, we overexpressed PTEN and PLK1 simultaneously to evaluate their expressions at the mRNA and molecular levels, respectively. Figure 5c-e reveals that after PLK1 overexpression, the PLK1 level increased significantly (P < 0.001), and the PTEN level decreased significantly (P < 0.001). After the simultaneous overexpression of PLK1 and PTEN, the PLK1 levels decreased significantly (P < 0.001), and those of PTEN increased significantly (P < 0.001). The same trend was observed at the mRNA level [Figure 5f and g].
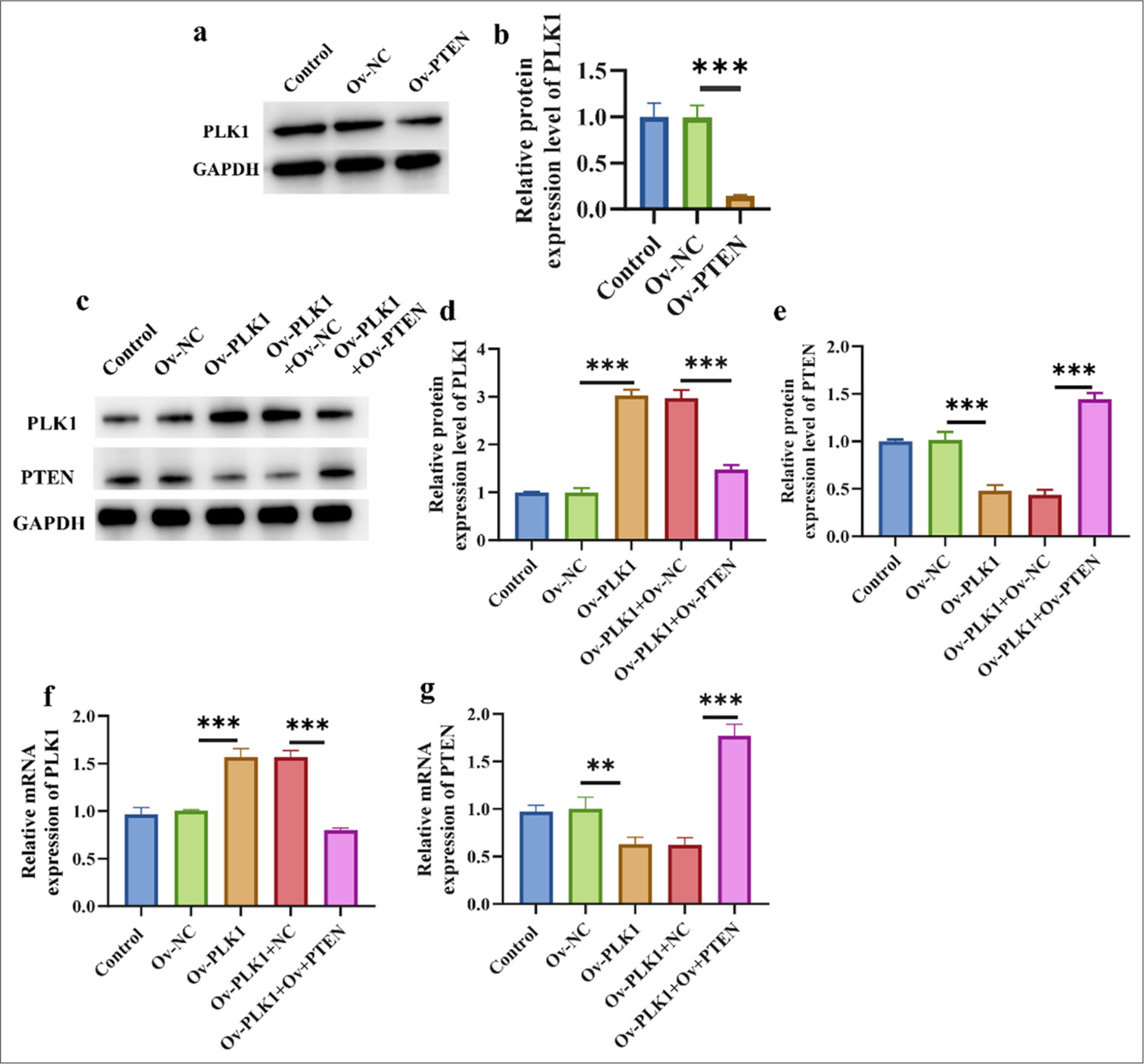
- Up-regulation of PTEN inhibited PLK1. (a and b) Effect of PTEN overexpression on PLK1 protein expression. (c-e) Western blot verified the efficiency of overexpressions of PLK1 and PTEN. (f and g) RT-qPCR verified the efficiency of overexpressions of PLK1 and PTEN. n = 3 ✶✶P < 0.01, ✶✶✶ P < 0.001. Ov-PLK1: PLK1 overexpression plasmid, Ov-PLK1+Ov-NC: negative control to overexpression plasmid, Ov-PLK1+ Ov-PTEN: PTEN and PLK1 overexpression plasmid, RT-qPCR: Real-time quantitative polymerase chain reaction.
PTEN overexpression reverses the oncogenic effect of the PLK1 gene
Compared with those in the Ov-PLK1 + Ov-NC group, these protein levels were higher in the Ov-PLK1 + Ov-PTEN treatment group, which indicates that PTEN overexpression promotes autophagy in lung cancer cells (P < 0.01) [Figure 6a-d]. p62 protein levels were significantly lower in the Ov-PLK1+Ov-PTEN treatment group (P < 0.001) [Figure 6e]. Electron microscopy results disclose that PLK1 overexpression reduced the formation of autophagosomes, and that of PTEN promoted their formation. This finding suggests that PTEN overexpression can promote the accumulation of autophagosomes, which results in the enhanced autophagy process in lung cancer cells [Figure 6f]. Detection of intracellular ROS levels showed the following changes in various treatment groups: Ov-PLK1 + OvPTEN > Ov-NC > Ov-PLK1. This result means that PTEN overexpression can affect intracellular ROS levels, and PLK1 overexpression reverses this effect (P < 0.05, and P < 0.001) [Figure 6g and h]. Cell viability assay serves as an important indicator for the evaluation of cell proliferation and survival capacity. The experimental findings demonstrate that the Ov-PLK1 group had greater cell viability, whereas cell viability decreased after cotransfection with Ov-PLK1 and Ov-PTEN (P < 0.001) [Figure 6i-l]. This result implies that PLK1 overexpression can reverse the inhibitory effect of PTEN upregulation on the clonogenic ability of lung cancer cells. In summary, PTEN upregulation can inhibit lung cancer cell proliferation and clonogenic potential by influencing intracellular ROS levels, which promotes the expression of autophagy-related proteins and aids in the accumulation of autophagosomes. These findings are supported by the experimental results mentioned above. However, PLK1 overexpression reverses these inhibitory effects.
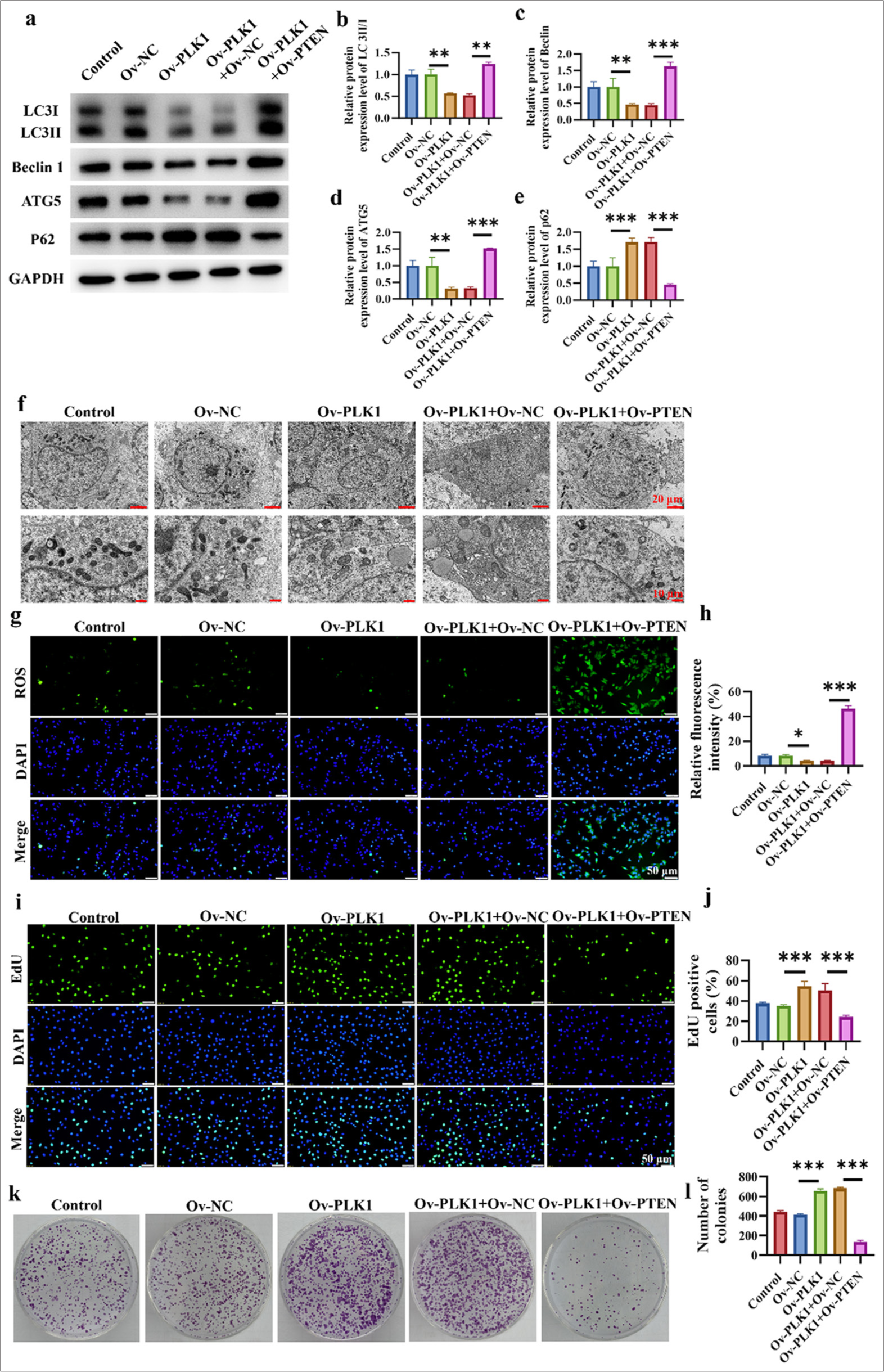
- PTEN overexpression counteracts the oncogenic effects of the PLK1 gene. (a-e) Expression of autophagy-related proteins in lung cancer cells in different treatment groups. (f) Promotion of autophagosome accumulation in lung cancer cells. (g and h) Detection of ROS levels in cells. (i and j) Cell viability assay of various treatment groups. (k and l) Clonogenic capacity assay of different treatment groups. n = 3 (✶P < 0.05, ✶✶ P < 0.01, ✶✶✶ P < 0.001).
Relationship between PLK1 and PTEN in xenograft tumor models
We further validated the results of in vitro experiments using a mouse xenograft tumor model. As illustrated in Figure 7a-c, the overexpression of PLK1 significantly promoted tumor growth (P < 0.01), but after the overexpressions of PLK1 and PTEN, tumor growth was significantly inhibited (P < 0.01). According to the pathological analysis in Figure 7d, after overexpression of PLK1, the cell space enlarged and exhibited an irregular arrangement. However, this situation was reversed after the overexpression of PTEN, similar to the tumor section image in the NC group. Finally, the expression levels of autophagy-related proteins in tumor tissues were analyzed at the RNA and protein levels, respectively (P < 0.001) [Figure 7e-l]. After the overexpressions of PLK1 and PTEN, the protein and RNA levels of LC3, Beclin1, and ATG5 were significantly increased and that of P62 was significantly decreased.
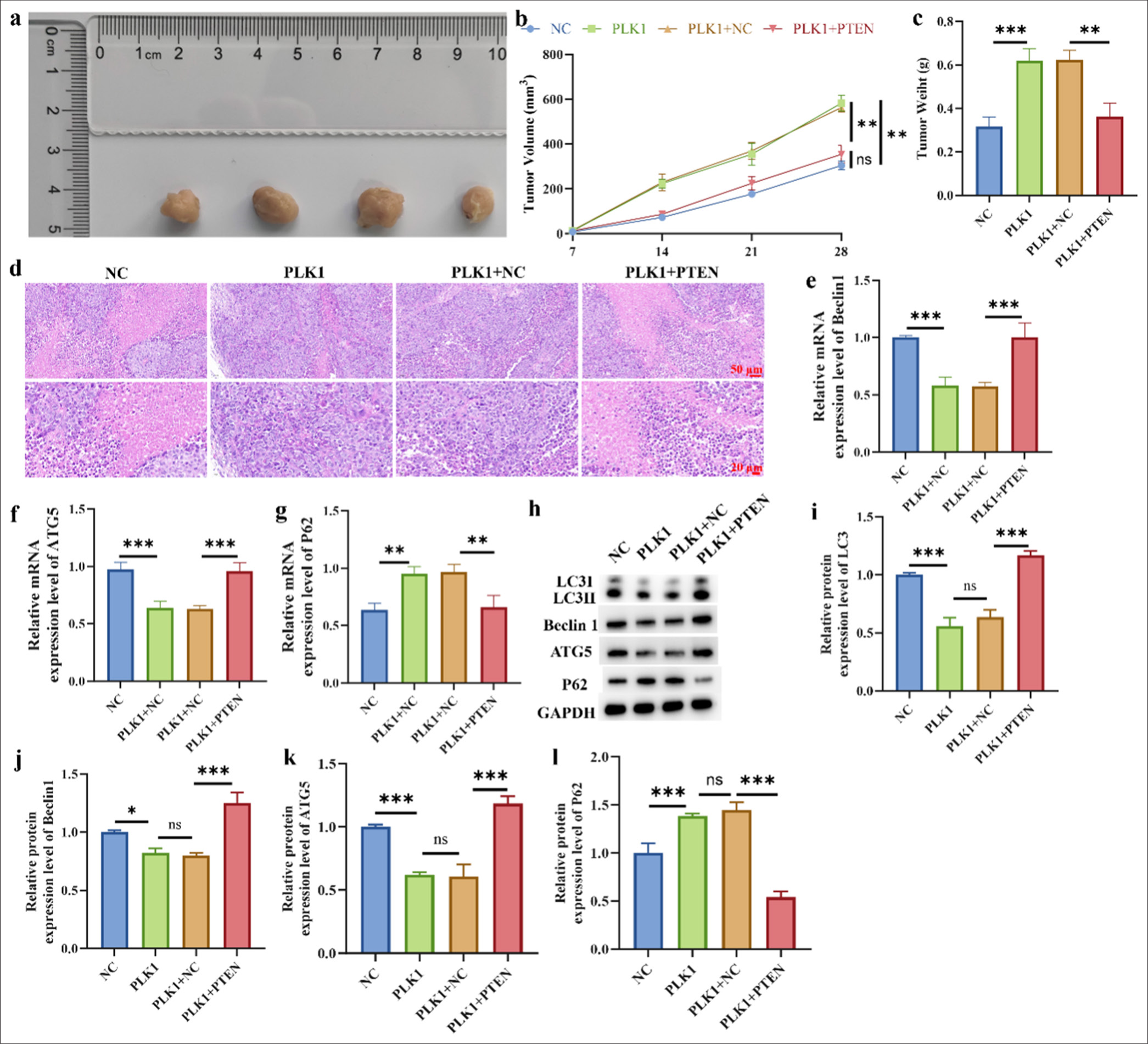
- Relationship between PLK1 and PTEN in xenograft tumor models. (a) A549 cell xenoinhibitory tumor representation. (b and c) Tumor weight and volume. (d) H&E staining of tumor tissue. (e-g) mRNA level of Beclin1, AGT5, and P62. (h-l) Protein level of LC3, Beclin1, AGT5 and P62. n = 3 (ns: No statistical difference, ✶P < 0.05, ✶✶ P < 0.01, ✶✶✶P < 0.001). H&E: Hematoxylin and eosin, LC3: Light chain 3, ATG5: Autophagy protein 5.
DISCUSSION
As one of the diseases with the highest global cancer mortality rates, the improvement of treatment strategies and the development of novel therapeutic drugs for lung cancer are clinically important.[21] To investigate the possible processes by which PTEN inhibits lung cancer, this study concentrated on examining how PTEN reduces lung cancer cell growth by upregulating autophagy and inhibiting the oncogene PLK1. Our results demonstrate that PTEN effectively inhibited the proliferation of lung cancer cells, and this inhibitory effect may be related to its downregulation of PLK1 expression and upregulation of autophagy.
PLK1, as an important cell cycle regulatory protein, contributes to the development of various tumors.[12,22] The overexpression of PLK1 can promote tumor cell proliferation, and the inhibition of PLK1 activity suppresses tumor growth.[23] Our experimental results show that PTEN can reduce the protein levels of PLK1, which suggests that PTEN may exert its antitumor effects by inhibiting the expression of PLK1. PLK1 is a well-known tumor promoter whose level is increased in a variety of cancers and is essential for controlling the cell cycle. In line with the findings of our study, PLK1 silencing inhibits the growth of lung cancer cells. Our data further support the importance of PLK1 as a potential target for lung cancer therapy.
Autophagy, as an intracellular degradation pathway, plays an important role in maintaining cellular homeostasis and responding to cellular stress. Recent studies have also revealed the complex role of autophagy in tumor formation. According to our research, PTEN can upregulate the expressions of proteins related to autophagy, and the use of autophagy inhibitors confirmed autophagy enables PTEN to prevent the growth of lung cancer cells. The findings imply that PTEN may stop lung cancer cell proliferation by initiating the autophagy pathway. We observed that PTEN can inhibit the proliferation of lung cancer cells, which is consistent with the results of other groups who reported the antiproliferative effects of PTEN in other cancer models.[24] In addition, we observed that PTEN exerts its inhibitory effect by downregulating PLK1, whose high expression in many tumor types is associated with a poor clinical prognosis. According to Ruf et al., PLK1 and mammalian target of rapamycin complex 1 (mTORC1) colocalize in lysosomes. The overexpression of PLK1 inhibits mTORC1 and actively promotes autophagy, whereas its suppression through shRNA or medication treatment increases the mammalian target of rapamycin lysosomal localization and decreases autophagy.[25] Moreover, we discovered that PTEN can increase the expression of proteins linked to autophagy, which is consistent with the findings of Xu et al., who discovered that autophagy contributes to tumor growth prevention.[26] In the clinical trial of Ferrara et al., the absence of PTEN in patients resulted in poor outcomes for patients. In addition, it promoted the PI3K/AKT pathway, which led to cell proliferation and growth.[27] Vidotto et al. also proved that PTEN serves as an important target of lung cancer tumor immune response.[28] However, our work is the first to reveal a new mechanism in which PTEN synergistically inhibits the proliferation of lung cancer cells by inhibiting PLK1 and promoting autophagy.
This study has achieved important academic achievements by elucidating the potential molecular mechanisms underlying the inhibition of lung cancer cell proliferation by PTEN. We not only confirmed that PTEN can suppress the expression of PLK1 but also promote autophagy activation in lung cancer cells. These results may provide fresh perspectives on lung cancer treatment and are crucial for comprehending PTEN’s antitumor actions. Furthermore, this work offers fresh proof of PLK1’s potential as a therapeutic target in lung cancer management and may aid in the development of innovative antitumor medications. Song et al. observed an increased propensity to metastasis, decreased PTEN levels, and increased pAkt activity in gemcitabine-resistant HPAC and PANC-1 cell models as indicative features.[29] This condition also provides a direction for our follow-up research.
However, this study encountered several limitations. The regulatory mechanism of PTEN on PLK1 expression remains unclear and thus requires further molecular biological experiments to elucidate its specific regulatory pathways. Furthermore, whether PTEN can produce synergistic effects with existing lung cancer treatment, drugs must be investigated to provide effective treatment options for clinical therapy.
SUMMARY
Our study preliminarily revealed the potential mechanism by which PTEN inhibits lung cancer cell proliferation through the inhibition of PLK1 and the upregulation of autophagy. These findings provide scientific evidence for the application of PTEN in lung cancer treatment and lay the foundation for further research. Future research will delve deeper into the exploration of the antitumor effects of PTEN and its potential clinical applications.
ACKNOWLEDGMENT
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All the data used for analysis in this study are supported by existing data in the network database, and in this published article, all the study’s generated data are presented.
ABBREVIATIONS
AKT: Protein kinase B
ATG5: autophagy related 5
FBS: Fetal bovine serum
H&E: Hematoxylin and Eosin
p62: Sequestosome 1
PI3K: Phosphoinositide 3-kinase
PLK1: Polo-like kinase 1
PTEN: Phosphatase and tensin homolog deleted on chromosome ten
RT-qPCR: Real-time quantitative polymerase chain reaction
TEM: Transmission electron microscopy
AUTHOR CONTRIBUTIONS
LMH: Designed the research study; LMH, WZJ, and PW: Performed the research; LMH: Collected and analyzed the data; WZJ and PW: Involved in drafting the manuscript and all authors have been involved in revising it critically for important intellectual content. All authors give final approval of the version to be published. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study has been approved by the ethics committee of Weifang Traditional Chinese Medicine Hospital, approval No. 2024YX122, dated 2024.07.31. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Weifang Traditional Chinese Medicine Hospital.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
How to cite this article: Jiang W, Wang P, Huang L. Upregulation of phosphatase and tensin homolog deleted on chromosome ten inhibits lung cancer cell proliferation by suppressing the oncogene polo-like kinase 1 and inducing autophagy. CytoJournal. 2025;22:10. doi: 10.25259/Cytojournal_146_2024
HTML of this article is available FREE at: https://dx.doi.org/10.25259/Cytojournal_146_2024
References
- Cancer progress and priorities: Lung cancer. Cancer Epidemiol Biomarkers Prev. 2019;28:1563-79.
- [CrossRef] [PubMed] [Google Scholar]
- Current and future development in lung cancer diagnosis. Int J Mol Sci. 2021;22:8661.
- [CrossRef] [PubMed] [Google Scholar]
- Lung cancer 2020: Epidemiology, etiology, and prevention. Clin Chest Med. 2020;41:1-24.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology of lung cancer In: Lung Cancer Imaging. United Kingdom: IOP Publishing Ltd; 2019. p. :4-1-4-15.
- [CrossRef] [Google Scholar]
- PTEN: Tumor suppressor and metabolic regulator. Front Endocrinol (Lausanne). 2018;9:338.
- [CrossRef] [PubMed] [Google Scholar]
- PTEN, PTENP1, microRNAs, and ceRNA networks: Precision targeting in cancer therapeutics. Cancers (Basel). 2023;15:4954.
- [CrossRef] [PubMed] [Google Scholar]
- Therapeutic implications of PTEN in non-small cell lung cancer. Pharmaceutics. 2023;15:2090.
- [CrossRef] [PubMed] [Google Scholar]
- PLK1 inhibition-based combination therapies for cancer management. Transl Oncol. 2022;16:101332.
- [CrossRef] [PubMed] [Google Scholar]
- Present and future perspective on PLK1 inhibition in cancer treatment. Front Oncol. 2022;12:903016.
- [CrossRef] [PubMed] [Google Scholar]
- Development of a nanoparticle-based immunotherapy targeting PD-L1 and PLK1 for lung cancer treatment. Nat Commun. 2022;13:4261.
- [CrossRef] [PubMed] [Google Scholar]
- Inhibition of the PLK1-coupled cell cycle machinery overcomes resistance to oxaliplatin in colorectal cancer. Adv Sci. 2021;8:2100759.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting Plk1 sensitizes pancreatic cancer to immune checkpoint therapy. Cancer Res. 2022;82:3532-48.
- [CrossRef] [PubMed] [Google Scholar]
- PLK1 inhibition sensitizes breast cancer cells to radiation via suppressing autophagy. Int J Radiat Oncol Biol Phys. 2021;110:1234-47.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 2019;9:1167-81.
- [CrossRef] [PubMed] [Google Scholar]
- Autophagy as a therapeutic target in pancreatic cancer. Br J Cancer. 2021;124:333-44.
- [CrossRef] [PubMed] [Google Scholar]
- The role of autophagy in resistance to targeted therapies. Cancer Treat Rev. 2020;88:102043.
- [CrossRef] [PubMed] [Google Scholar]
- FBXO32-mediated degradation of PTEN promotes lung adenocarcinoma progression. Cell Death Dis. 2024;15:282.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol. 2019;12:134.
- [CrossRef] [PubMed] [Google Scholar]
- Active PLK1-driven metastasis is amplified by TGF-β signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2020;39:767-85.
- [CrossRef] [PubMed] [Google Scholar]
- PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett. 2019;467:9-18.
- [CrossRef] [PubMed] [Google Scholar]
- Effect of Astragalus polysaccharide on the expression of VEGF and EGFR in mice with Lewis transplantable lung cancer. J Coll Physicians Surg Pak. 2019;29:392-94.
- [CrossRef] [PubMed] [Google Scholar]
- PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy. 2017;13:486-505.
- [CrossRef] [PubMed] [Google Scholar]
- YAP manipulates proliferation via PTEN/AKT/mTOR-mediated autophagy in lung adenocarcinomas. Cancer Cell International. 2021;21:30.
- [CrossRef] [PubMed] [Google Scholar]
- PTEN loss as a predictor of tumor heterogeneity and poor prognosis in patients with EGFR-mutant advanced non-small-cell lung cancer receiving tyrosine kinase inhibitors. Clin Lung Cancer. 2021;22:351-60.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging role of PTEN loss in evasion of the immune response to tumours. Br J Cancer. 2020;122:1732-43.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-21 upregulation induced by promoter zone histone acetylation is associated with chemoresistance to gemcitabine and enhanced malignancy of pancreatic cancer cells. Asian Pac J Cancer Prev. 2013;14:7529-36.
- [CrossRef] [PubMed] [Google Scholar]








