Translate this page into:
Platelet-derived growth factor subunit B overexpression promotes lung cancer tumor growth and metastasis: The role of glucose metabolism

*Corresponding author: Gaofeng Qiao, Department of Thoracic Surgery, Shandong Public Health Clinical Center, Shandong University, Jinan City, Shandong, China. gaofengqiao@163.com
-
Received: ,
Accepted: ,
How to cite this article: Feng K, Cai X, Qiao G. Platelet-derived growth factor subunit B overexpression promotes lung cancer tumor growth and metastasis: The role of glucose metabolism. CytoJournal. 2025;22:33. doi: 10.25259/Cytojournal_190_2024
Abstract
Objective
Lung cancer represents a formidable global health challenge due to its substantial prevalence and mortality rates. Metabolic reprogramming, especially the transition to aerobic glycolysis, plays a pivotal role in the progression of lung cancer by sustaining the energy demands for rapid tumor proliferation. The prominent involvement of platelet-derived growth factor subunit B (PDGFB) in promoting the growth and metastasis of lung cancer through specific signaling cascades is well established in. Nonetheless, further research is imperative to elucidate the intricate regulatory mechanisms of PDGFB in glucose metabolism and its implications for the advancement of lung cancer. Our study is dedicated to exploring the effect of PDGFB on lung cancer by modulating glucose metabolism.
Material and Methods
First, we determined the expression patterns of PDGFB in various lung cancer cell lines (A549, H460, HCC827, and H1975) using quantitative real-time polymerase chain reaction and Western blot analyses. We measured the expression levels of PDGFB and Ki-67 in tumor tissues from lung cancer patients through immunohistochemistry. We then transfected lung cancer cells with a PDGFB overexpression (PDGFB OE) plasmid. The effects of PDGFB OE and galactose + PDGFB OE co-treatment on cell migration and invasion characteristics were assessed using wound healing and Transwell assays. The impact of PDGFB OE and galactose + PDGFB OE co-treatment on the proliferation capacity of lung cancer cells was evaluated through colony formation and 5-ethynyl-2’-deoxyuridine staining assays. We also measured the effects of PDGFB OE on mitochondrial function and glycolytic capacity in lung cancer cells using extracellular acidification rate assay (ECAR) measurement methods.
Results
Elevated levels of PDGFB expression were markedly detected in various lung cancer cell lines, notably A549 and H460 (P < 0.001). This observation was validated by the analysis of tumor samples from patients with lung cancer who exhibited heightened PDGFB expression in tumor tissues (P < 0.001). Moreover, an association was found between increased levels of Ki67 expression and elevated PDGFB expression (P < 0.001). The upregulation of PDGFB was linked to heightened migratory (P < 0.001), invasive (P < 0.001), and proliferative (P < 0.001) capacities of the cells. Furthermore, an elevation in lactate levels and ECAR (P < 0.001) was noted in the PDGFB OE group, along with increased levels of glycolysis-related regulatory proteins. The inhibition of aerobic glycolysis with galactose effectively mitigated the PDGFB-induced enhancement of lung cancer cell proliferation and migration.
Conclusion
By affecting glucose metabolism, PDGFB drives the growth and metastasis of lung cancer, underscoring its potential as a promising therapeutic target for the management of this complex disease.
Keywords
Cell proliferation
Glycolysis
Lung cancer
INTRODUCTION
Lung cancer poses a significant global health challenge as one of the most prevalent cancers worldwide, with both its incidence and mortality rates raising concerns.[1] A distinctive feature of tumor cells, metabolic reprogramming plays a crucial role in the growth and spread of cancer, particularly in lung cancer.[2] Among the metabolic pathways, aerobic glycolysis stands out as a key player in lung cancer progression.[3] Tumor cells rely on aerobic glycolysis to generate abundant lactate and ATP, adapting to their rapid proliferation and heightened metabolic demands, a phenomenon known as the “Warburg effect.” Tumor cells that heavily rely on this metabolic trait often exhibit heightened glucose uptake and rewiring of glucose metabolic pathways.[4]
Past studies illuminated platelet-derived growth factor subunit B (PDGFB) as a pivotal cytokine that fuels tumor growth and metastasis across various malignancies.[5,6] In the case of lung cancer, the expression levels of PDGFB closely correlate with tumor development and prognosis.[6,7] By binding to its receptor, PDGFB triggers signaling pathways that boost the proliferation, invasion, and migration of tumor cells.[8] Furthermore, within lung cancer research, PDGFB has been observed to play a role in modulating the tumor microenvironment, fostering tumor angiogenesis, and thereby influencing the progression of lung cancer.[9] While some headway has been made in understanding how PDGFB regulates lung cancer cells, numerous questions persist, beckoning exploration. Existing literature underscores that PDGFB influences lung cancer cell proliferation and migration by activating specific signaling pathways.[10] In addition, PDGFRβ has been reported to be involved in the process of glucose metabolism.[11,12]
The intricate regulatory mechanisms of PDGFB in this process, its crosstalk with other metabolic pathways, and how it impacts the growth and spread of lung cancer necessitate further scrutiny. In our present study, we devised experimental lung cancer cell lines overexpressing PDGFB alongside a blank control group.
Through diverse methods encompassing cell morphology experiments, gene expression analyses, and immunohistochemical staining, we delved deeply into the pivotal role of PDGFB in the evolution of lung cancer. In particular, we scrutinized genes related to glucose metabolism in lung cancer cell lines to unveil the regulatory effect of PDGFB on glucose metabolism.
By dissecting the mechanisms through which PDGFB influences lung cancer development and its role in reshaping the glucose metabolism of lung cancer cells, we aim to enhance our comprehension of lung cancer’s metabolic traits and the crucial regulatory mechanisms underpinning tumor cell growth. This endeavor not only aids in unraveling the molecular underpinnings of lung cancer progression but also furnishes valuable insights for identifying novel therapeutic targets in the battle against cancer.
MATERIAL AND METHODS
Cells, tissues, and reagents
A549 (BFN60800665), H460 (BFN60800670), HCC827 (BFN60800668), and H1975 (BFN608006102) cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). These specific cell lines were cultivated in Dulbecco’s modified eagle medium (DMEM, 11965092, Gibco™, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (11965092, Gibco™, Grand Island, NY, USA) to facilitate their growth and upkeep. The cells were housed in 37°C incubator with 5% CO2. What’s more, depending on the experiment, DMEM was further supplemented with 10 Mm glucose (A2494001, Thermo Fischer Scientific, Waltham, Ma, USA) or 10 mM galactose (15572-79-9, MACKLIN, Shanghai, China). Before the commencement of the experiments, screenings were performed on the cell lines for mycoplasma contamination, and their authenticity was validated through short-tandem repeat analysis.
Tumor specimens were collected from 12 patients with lung cancer and stored corresponding non-cancerous tissues at -80°C. All cases are newly diagnosed and have not undergone chemotherapy. The study obtained ethical approval, and informed consent was obtained from all participants. The clinical data of lung cancer patients involved in this study are shown in Table 1.
| Groups | Proportion (%) |
|---|---|
| Gender | |
| Male | 6 (50) |
| Female | 6 (50) |
| Ages (years) | |
| 31–40 | 2 (16.7) |
| 41–50 | 3 (25) |
| 51–60 | 3 (25) |
| 61–70 | 2 (16.7) |
| >70 | 2 (16.7) |
| Clinical lung cancer staging | |
| I stage | 3 (25) |
| II stage | 4 (33.33) |
| III stage | 3 (25) |
| IV stage | 2 (16.7) |
| Family history | |
| Yes | 4 (33.33) |
| No | 8 (66.67) |
| Religion | |
| Yes | 6 (50) |
| No | 6 (50) |
| Level of education | |
| Bachelor degree or above | 4 (33.33) |
| Junior college | 3 (25) |
| High school | 3 (25) |
| Junior high school | 1 (8.33) |
| Primary school and below | 1 (8.33) |
| Place of residence | |
| Cities | 6 (50) |
| Rural areas | 6 (50) |
In the realm of DNA manipulation, a set of restriction endonucleases (Lot: 3159, Takara, Tokyo, Japan), including HindIII, EcoRI, SacI, and XhoI were utilized, alongside the T4 DNA ligase (2011A, Takara, Tokyo, Japan) which aided in the merging of DNA fragments. In addition, polymerase chain reaction (PCR) and reverse transcription experiments were carried out with the employment of SYBR Premix Ex Taq™ II (RR390Q, Takara, Tokyo, Japan) and reverse transcription tools, particularly the PrimeScript™ RT reagent kit (RR037Q, Takara, Tokyo, Japan).
The study also made use of various other materials such as the plasmid extraction and gel recovery kits (12123, QIAGEN, Duesseldorf, Germany), RNA extraction reagent Trizol (15596018CN, Invitrogen, Carlsbad, CA, USA) for transfection and RNA extraction procedures, and the RIP and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel preparation kits (P0012AC, Beyotime, Beijing, China) for RNA immunoprecipitation and protein analysis experiments. In addition, materials such as polyvinylidene fluoride (PVDF) membrane and protein detection reagents (08321S, Merck Millipore, Billerica, MA, USA) were used to facilitate protein transfer and detection processes in the study.
Ethics and informed consent
This study has been approved by the Medical Ethics Committee of Shandong Public Health Clinical Center, Shandong University (approval No. KYLL-20230617), and informed consent was obtained from all clinical participants. It complies with the Declaration of Helsinki.[13]
Cell transfection
In brief, 1 × 105 cells were inoculated in 24-well plates of 500 uL medium and cultured overnight at 37°C with 5% CO2. When the cell density was 40–50%, transfection was performed, and the fresh medium was replaced 24 h later. In the study, A549 and H460 cells were categorized into the PDGFB overexpression (PDGFB OE) group and the control group. Lipofectamine 2000 (18501, Invitrogen, Carlsbad, CA, USA) was used as the transfection agent. Specific plasmids were designed for the overexpression of PDGFB and OENC (pCMV-negative control). The PDGFB OE sequence was synthetically crafted by GeneChem (Shanghai, China), with the forward sequence as 5’- AATTCAGCTTCCCT GGCTCTAGGCGAGTCGACATATTGGCCTA-3’ and the reverse as 5’-CTAACTTGTCGACTCTCCAGAAC GGTG TGCTTTGTGAG-3’. Subsequent procedures included polyadenylation of RNA using poly(A) polymerase (0AS72, American Research Products, Grandville, MI, USA), followed by transfection into the cells. Post-transfection, cells were nurtured to ensure effective integration of transfection vectors and gene activation, with analysis conducted through Western blotting for transfection assessment.
Wound healing cell migration assay
To conduct a scratch assay, 2 × 104 A549 or H460 cells were seeded in 6-well plates with culture medium until they reached 90% confluence. Subsequently, uniform scratches were created on the cell monolayer using a plastic pipette tip to simulate the process of wound healing. Afterward, the cells were rinsed with a pre-warmed culture medium containing fetal bovine serum to eliminate any cell debris. A fresh culture medium supplemented with fetal bovine serum was then added, and the cells were cultured further. Images of the scratches are captured at 0 and 24-h time points using an Olympus light microscope (CX23, Olympus, Tokyo, Japan). The relative migration distance of the cells is measured using ImageJ software (v1.8.0.345, National Institutes of Health, Bethesda, MD, USA) to evaluate migration ability. Experimental and control groups were established for comparison, and the results were analyzed by examining image data and migration distances to draw meaningful conclusions.
Transwell assay
Transwell chambers (3401, Corning Incorporated, NY, USA) were utilized for the experiment. Initially, A549 and H460 cells underwent a 24 h treatment of serum deprivation in a culture medium. Subsequently, the cells were replenished with serum-free medium and placed in a 37°C incubator for rehydration. The cell density in the upper chamber was adjusted according to the initial seeding density. Following this, the 24-well plate was placed in a 37°C incubator with 5% CO2 and 90% humidity for a duration of 12 h. The cells were then fixed for 30 min, stained with crystal violet solution (C0121, Beyotime, Shanghai, China) for 20 min, and observed under a microscope (CX23, Olympus, Tokyo, Japan).
Matrigel invasion assay
For evaluating cell invasion, the Invasion Chamber coated with Matrigel from (619203, BD Biosciences UJ, USA) was employed. In short, 2 × 104 A549 and H460 cells were cultured in the upper chamber of the Transwell insert using serum-free medium for 72 h. The cells that penetrated were fixed using 4% paraformaldehyde and then stained with 0.5% crystal violet for 20 min. Subsequently, the count of cells that had penetrated in each group was conducted under a light microscope (CX23, Olympus, Tokyo, Japan).
Clonogenic assay
During the clonogenic assay, A549 and H460 cells were plated in a 24-well plate with precision and care. Cells were digested with trypsin, centrifuged to obtain cell suspension, and continued to be cultured for 10 days at a concentration of 1000/well. After the incubation period, the cells were securely fixed using a 4% paraformaldehyde solution. Following fixation, the cells were treated with a crystal violet staining solution to enable visualization of their characteristics. Subsequently, the samples were carefully examined under a microscope (CX23, Olympus, Tokyo, Japan) to observe and analyze the formation of clonal structures.
5-ethynyl-2’-deoxyuridine (EdU) incorporation
The kit for EdU incorporation (C0081, Beyotime, Beijing, China) was utilized to examine the proliferation in A549 and H460 cells. Initially, the cells were treated with the EdU working solution, washed with phosphate-buffered saline, and subsequently stained with 4’,6-diamidino-2-phenylindole (D1306, Invitrogen, Carlsbad, CA, USA) to visualize the cell nuclei. Following fixation, the cells were left to air-dry at room temperature and then examined using a fluorescence microscope (CX33, Olympus, Tokyo, Japan) to evaluate the proliferation in A549 and H460 cells. The cells showing EdU-positive results, indicating apoptotic cells, emitted red fluorescence, while the cell nuclei emitted blue fluorescence. Cell counting was conducted in four different fields, and the ratio of EdU-positive cells to the total cell count was documented as the proliferative cell ratio.
Ki67 Staining
The expression levels of Ki-67 in cells and tumor tissues were assessed through immunocytochemistry and immunohistochemistry (IHC). A549 and H460 cell slides were prepared, and the tissues were embedded with paraffin. The slides were then subjected to deparaffinization and rehydration procedures. Antigen retrieval was performed to reveal the epitopes. Following this, the primary Ki67 antibody (ab16667, Abcam, Cambridge, MA, USA) was allowed to incubate with the cell slides overnight. Subsequent to rinsing, an Anti-Rabbit IgG antibody (ab150077, Abcam, Cambridge, MA, USA) was applied to the slides. The slides were then washed again. Finally, the stained slides were examined under a microscope (CX23, Olympus, Tokyo, Japan) for visualization and analysis purposes. Cell counting was conducted in four different fields, and the ratio of Ki67-positive cells to the total cell count was documented as the proliferative cell ratio.
IHC analysis
IHC analysis was used to detect the expression of PDGFB protein in tissue samples. The process involves dehydration and rehydration of tissue sections, followed by treatment with hydrogen peroxide and hot citrate buffer. Subsequently, the primary antibody of PDGFB (ab107101, Abcam, Cambridge, MA, USA) is added to the sample for overnight incubation. The second antibody (ab207997, Abcam, Cambridge, MA, USA) was incubated at room temperature for 1h. Next, a diaminobenzidine kit (BL732A, BioSharp, Hefei, China) is used for staining, and the results are evaluated by microscope (BX46, Olympus Corporation, Tokyo, Japan) observation of staining intensity and the proportion of positive staining. Finally, the expression level of PDGFB in the sample is determined based on the staining intensity score. The following proportion scores were assigned to the sections: 1, 0–1%; 2, 2–10%; 3, 11–30%; 4, 31–70%; and 5, 71–100%. The staining intensity was rated on a scale of 0–3: 0, negative; 1, weak; 2, moderate; and 3, strong. Then, the proportion and intensity scores were combined to obtain a total score (range, 0–8).
Detection of extracellular acidification rate assay (ECAR) and ATP
The ECAR was measured on an XF pro Seahorse flux analyzer (Agilent Technologies, Santa Clara, CA). A549 and H460 cells were hydrated overnight in a 37℃ nonCO2 incubator with calibration buffer in a Seahorse 96-well plate at 6000 cells per well concentration. The ECAR was measured in real-time in an XFe96 analyzer using a Seahorse XF Glycolytic Stress Assay Kit (103020-100, Agilent, Santa Clara, CA, USA). ECAR measurements were performed under basal conditions, followed by the sequential addition of 1.5 mM oligomycin and 50 mM 2-deoxyglucose.
Adenosine triphosphate (ATP) production was determined with the ATP Determination Kit (Lot: A22066, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Briefly, cells with different treatments were homogenized in the lysis buffer. The results were determined by luminescence (Glomax 20/20 luminometer, Promega, Madison, Wisconsin, USA) and normalized to protein concentration.
Determination of glucose consumption and lactate production
Glucose and lactate detection kits (BC2505 and BC2235) purchased from Solarbio (Beijing, China) were used for the determination of glucose and lactate concentrations before and after 24 h cell culture in OC cells, following their manufacturer’s protocols. In addition, a pH meter (PB-11 Basic Meter, Sartorius, Gottingen, Germany) was used for the measurement of pH value in the cell culture medium.
Western blot analysis
Total protein was separated by SDS-PAGE gel electrophoresis and was subsequently transferred onto a PVDF membrane (IPVH00010, Millipore Corporation, Billerica, MA, USA). The membrane was then blocked with 5% skim milk at 37°C for 2 h. Specific antibodies (all purchased from Abcam, Cambridge, MA, USA), including PDGFB (ab178409, 1:400), HK2 (ab209847, 1:400), PKM2 (ab85555, 1:400), glucose transporter 1 (GLUT1) (ab115730, 1:400), LDHA (ab52488, 1:400), GAPDH (ab9485, 1:000) and β-actin (ab8227, 1:1000), were incubated with the membrane overnight. Subsequently, the corresponding horseradish peroxidase Anti-Rabbit IgG antibody (ab288151, Abcam, Cambridge, MA, USA) was incubated with the membrane at 37°C for 1 h. Protein bands on the membrane were visualized using ECL luminescent fluid (BL520b, Biosharp Life Science, Hefei, Anhui, China) and imaged using the Bio-Rad system (12011319, Bio-Rad, Hercules, CA, USA). The expression of each protein was evaluated using β-actin as an internal reference.
Qualitative real-time PCR (qRT-PCR)
Total RNA was extracted using a kit (DP419), and the RNA was reverse-transcribed into complementary DNA (cDNA) using Quant cDNA first strand synthesis kit (KR103). Reverse transcription of RNA into cDNA was conducted according to the manufacturer’s instructions. Kits are provided by Tiangen (Beijing, China). The resulting reaction solution was utilized for fluorescence quantitative PCR (qPCR). The primer sequences used for the qPCR analysis are listed in Table 2. qRT-PCR was performed with GAPDH as the internal reference. The 2-ΔΔCt method was employed for result analysis.
| Primer Name | Primer sequences (5’-3’) |
|---|---|
| PDGFB-F | CGCTAACATCAA ATGGGGTG |
| PDGFB-R | TTGCTGACAATCTTGAGGGAG |
| GAPDH-F | CGCTAACATCAAATGGGGTG |
| GAPDH-R | TTGCTGACAATCTTGAGGGAG |
PDGFB: Platelet-derived growth factor subunit B, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, RT-qPCR: Qualitative real-time polymerase chain reaction, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Statistical analysis
GraphPad Prism (Version 8.0; La Jolla, CA, USA) was used for statistical analysis and graphical representation of the results. Measurement data are presented as mean ± standard deviation. Comparison between two groups was performed using a t-test, while one-way analysis of variance (ANOVA) was used for comparisons involving multiple groups. The Tukey method was used for multiple post hoc testing in oneway ANOVA. A significance level of P < 0.05 was considered statistically significant.
RESULTS
PDGFB is upregulated in lung cancer cells, with its expression levels escalating with the progression of malignancy
We first compared the expression levels of PDGFB in various lung cancer cell lines, including A549, H460, HCC827, and H1975. These lung cancer cell lines exhibited significantly elevated levels of PDGFB expression, especially in A549 (P < 0.001) and H460 (P < 0.001) when compared to normal lung epithelial cells BEAS-2B [Figure 1a and b]. Furthermore, patient-derived lung cancer samples were examined using PDGFB immunohistochemical staining, revealing significantly elevated PDGFB expression levels (P < 0.001) in tumor tissues in comparison to adjacent non-tumor tissues [Figure 1c and d]. In conclusion, the findings indicate that PDGFB is highly expressed in lung cancer cells, and its expression levels correlate positively with the malignancy grade, suggesting a potential role of PDGFB in the progression of lung cancer.
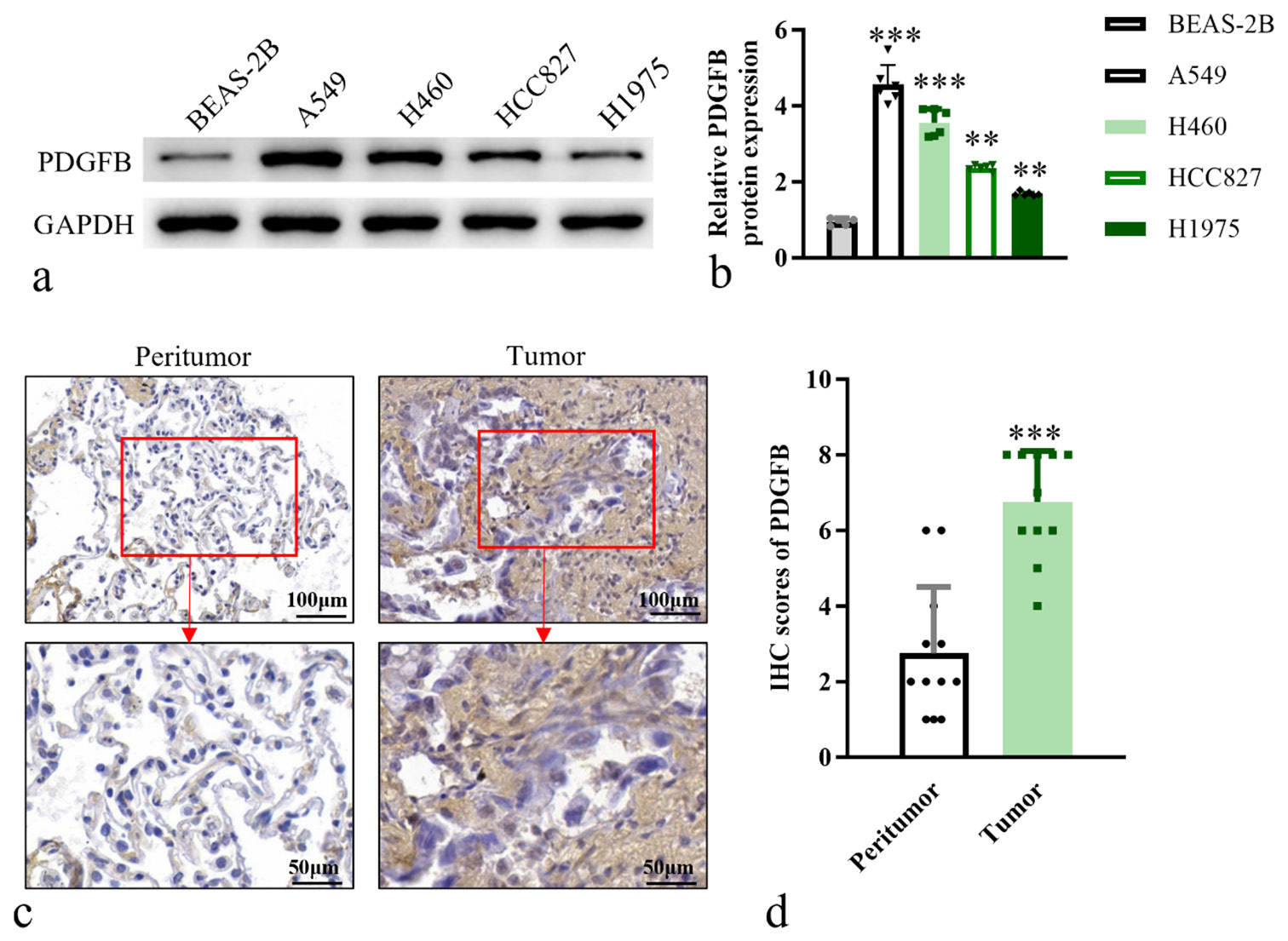
- PDGFB is upregulated in lung cancer cells, with its expression levels escalating with the progression of malignancy. (a) Western blot analysis of PDGFB in cell lines. (b) Histogram of relative PDGFB protein expression. (c) IHC analysis, objective: 200×, of PDGFB in tumor and adjacent normal tissue (n = 12) (d) Quantitative analysis of PDGFB expression levels in IHC staining. n = 6. ✶✶P < 0.01, ✶✶✶P < 0.001. Scale bar, 50 μm and 100 μm. PDGFB: Platelet-derived growth factor subunit B, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, IHC: Immunohistochemistry.
Overexpression of PDGFB in lung cancer cells led to enhanced migratory and invasive capabilities
Next, an experimental group overexpressing PDGFB was established through viral vector transfection to induce PDGFB OE in lung cancer cells, while a corresponding blank control group was concurrently set up. To validate the efficacy of PDGFB OE, western blotting and RT-PCR analyses were performed to assess the expression levels of PDGFB in the experimental (PDGFB OE) and control groups [Figure 2a-c].
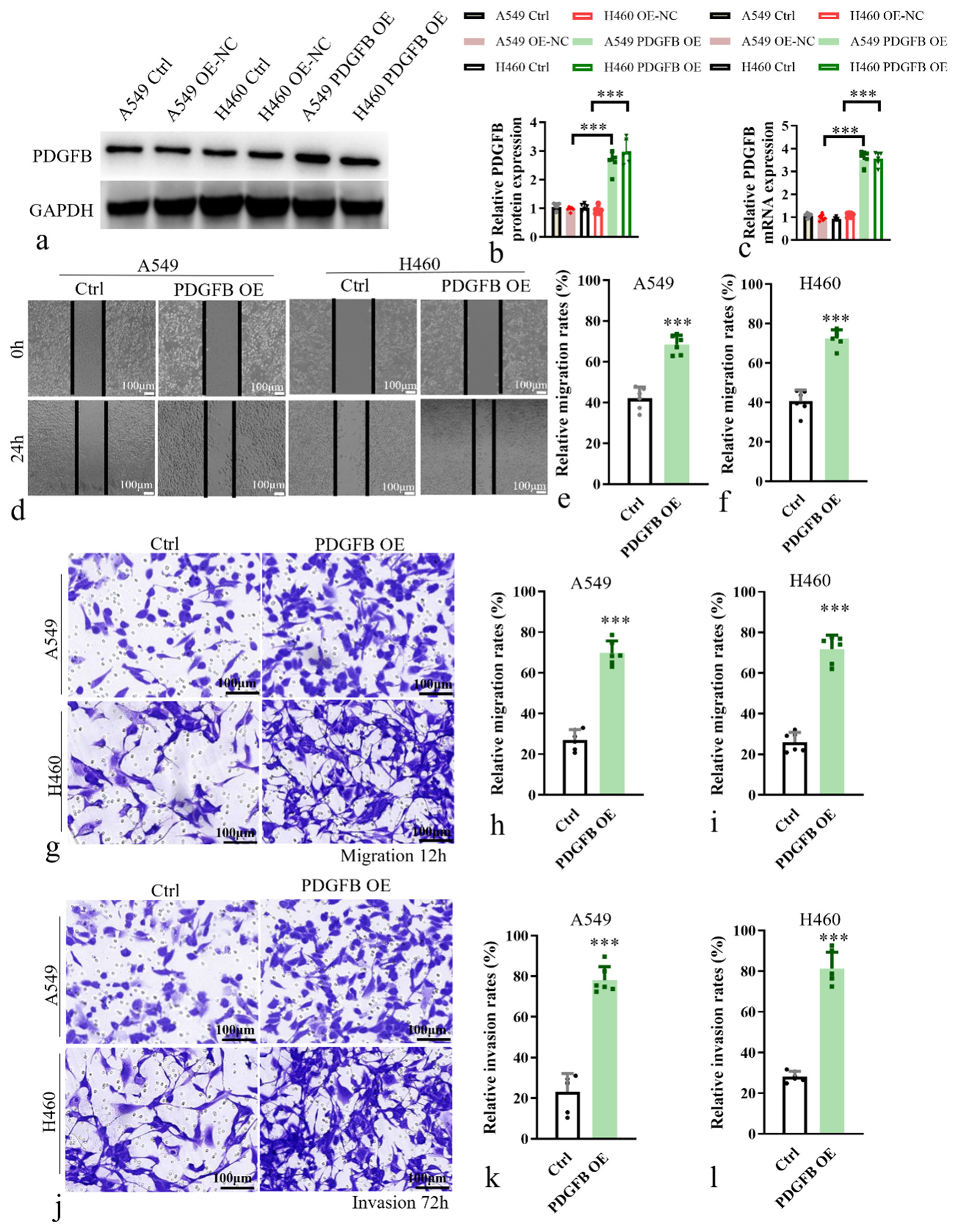
- Overexpression of PDGFB in lung cancer cells led to enhanced migratory and invasive capabilities. (a and b) Western blot analysis of PDGFB in overexpression cell lines. (c) Histogram of relative PDGFB mRNA expression. (d) Wound-healing cell migration assay in A549 and H460. (e and f) Relative migration rate in A549 and H460. (g-i) Transwell chamber assays in A549 and H460. (j-l) Matrigel-coated Transwell chamber assays in A549 and H460. Scale bar, 100 μm. n = 6. ✶✶✶P < 0.001. Ctrl: Control, PDGFB OE: Overexpression of PDGFB, PDGFB: Platelet-derived growth factor subunit B.
Functional assays were conducted to investigate the impact of PDGFB OE on cellular migratory and invasive properties. Scratch assays [Figure 2d-f] and Transwell chamber assays [Figure 2g-i] were employed to evaluate the migratory capacity of cells following PDGFB OE. It showed that the relative migration rate was a significant increase (P < 0.001) in the PDGFB OE group. In addition, Matrigel-coated Transwell chamber assays [Figure 2j-l] were utilized to assess the invasive potential of cells overexpressing PDGFB. Compared to the control group, the PDGFB OE group revealed a significant enhancement (P < 0.001) in the invasive abilities of A549 and H460 cell lines. These results revealed a significant enhancement in the migratory and invasive abilities of lung cancer cells in response to PDGFB OE. These findings underscore the potential role of PDGFB in promoting tumor cell migration and invasion, highlighting its implication in the metastatic behavior of lung cancer cells.
Impact of PDGFB OE on the proliferation ability of lung cancer cells
Next, we investigated the effects of PDGFB OE on the proliferation ability of human lung cancer cell lines A549 and H460. To assess cell proliferation, clonogenic assays were performed. A549 and H460 cells from the experimental (PDGFB OE) and control groups were plated at low density and allowed to form colonies. The number and size of colonies in the PDGFB OE group were significantly increased compared with the control group (P < 0.001) [Figure 3a and b]. Meanwhile, EdU incorporation was performed to detect actively proliferating cells. Cells were exposed to EdU, which is incorporated into actively dividing cells during DNA synthesis. The levels of EdU incorporation in the PDGFB OE group were significantly increased compared with the control group in A549 (P < 0.001) and H460 (P < 0.001) [Figure 3c-e]. As a marker of cell proliferation, staining using Ki67 antibody was conducted to determine the proportion of cells in the proliferative phase. The Ki67-positive cells were quantified and showed that the PDGFB OE group has more Ki67-positive cells than the control group (P < 0.001) [Figure 3f-h].
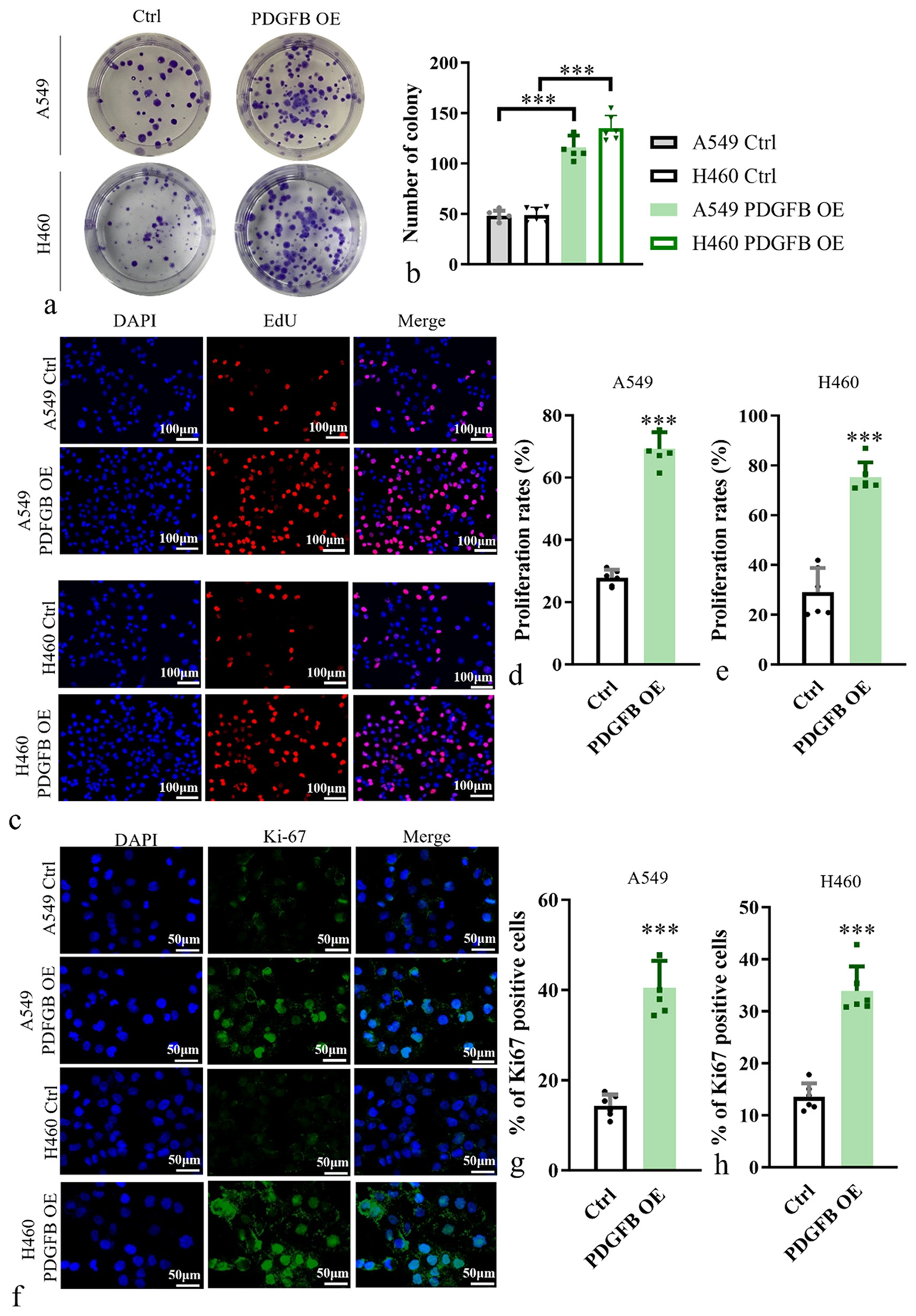
- Impact of PDGFB overexpression on the proliferation ability of lung cancer cells. (a and b) Analysis of proliferation ability of A549 and H460. (c-e) EdU incorporation assay, objective: 200×, was performed in A549 and H460. Scale bars, 100 μm. (f-h) Staining of Ki67 antibody in A549 and H460, objective: 200×. Scale bars, 50 μm. n = 6. ✶✶✶P < 0.001. EdU: 5-ethynyl-2’-deoxyuridine, PDGFB: Platelet-derived growth factor subunit B.
The results conclusively showed that PDGFB OE significantly enhanced the proliferation ability of lung cancer cells. This finding strongly suggests that PDGFB plays a crucial role in promoting lung cancer cell proliferation.
PDGFB modulates aerobic glycolysis of lung cancer cells
Research suggests that phosphoinositide 3-kinase/protein kinase B/mammalian target of the rapamycin signaling pathway is downstream of PDGFB and plays an essential part in provoking the Warburg effect in tumor cells and then increasing pyruvate dehydrogenase kinase 1 expression, which suppresses pyruvate dehydrogenase activity and shifts metabolism to aerobic glycolysis.[12]
Therefore, we conducted aerobic glycolysis assays on A549 and H460 cell lines in the PDGFB OE and control group. Evaluation of mitochondrial function, glycolysis, and oxidative phosphorylation processes was performed using ECAR measurements. Results revealed a significant increase in ECAR levels (P < 0.05) at various time points for the PDGFB OE group in A549 [Figure 4a] and H460 [Figure 4b]. Remarkably, a pronounced increase in lactate levels (P < 0.001) [Figure 4c and d] and glucose uptake (P < 0.001) [Figure 4e and f] were found to be elevated in the PDGFB OE group, signifying an upregulated glycolytic state. Meanwhile, a notable reduction was observed in both ATP levels (P < 0.001) on PDGFB OE [Figure 4g and h]. In addition, we assessed the PH value in the medium and found that the PDGFB OE group has a higher PH value in the medium [Figure 4i and j]. Furthermore, we assessed the expression of aerobic glycolysis biomarkers, hexokinase 2, pyruvate kinase M2, GLUT1, and lactate dehydrogenase A. Western blot analysis unveiled elevated levels of glycolysis-related regulatory proteins in the PDGFB OE group (P < 0.001) [Figure 4k-o]. Taken together, these findings substantiate PDGFB’s potential to modulate aerobic glycolysis, thereby impacting the functionality of lung cancer cells.
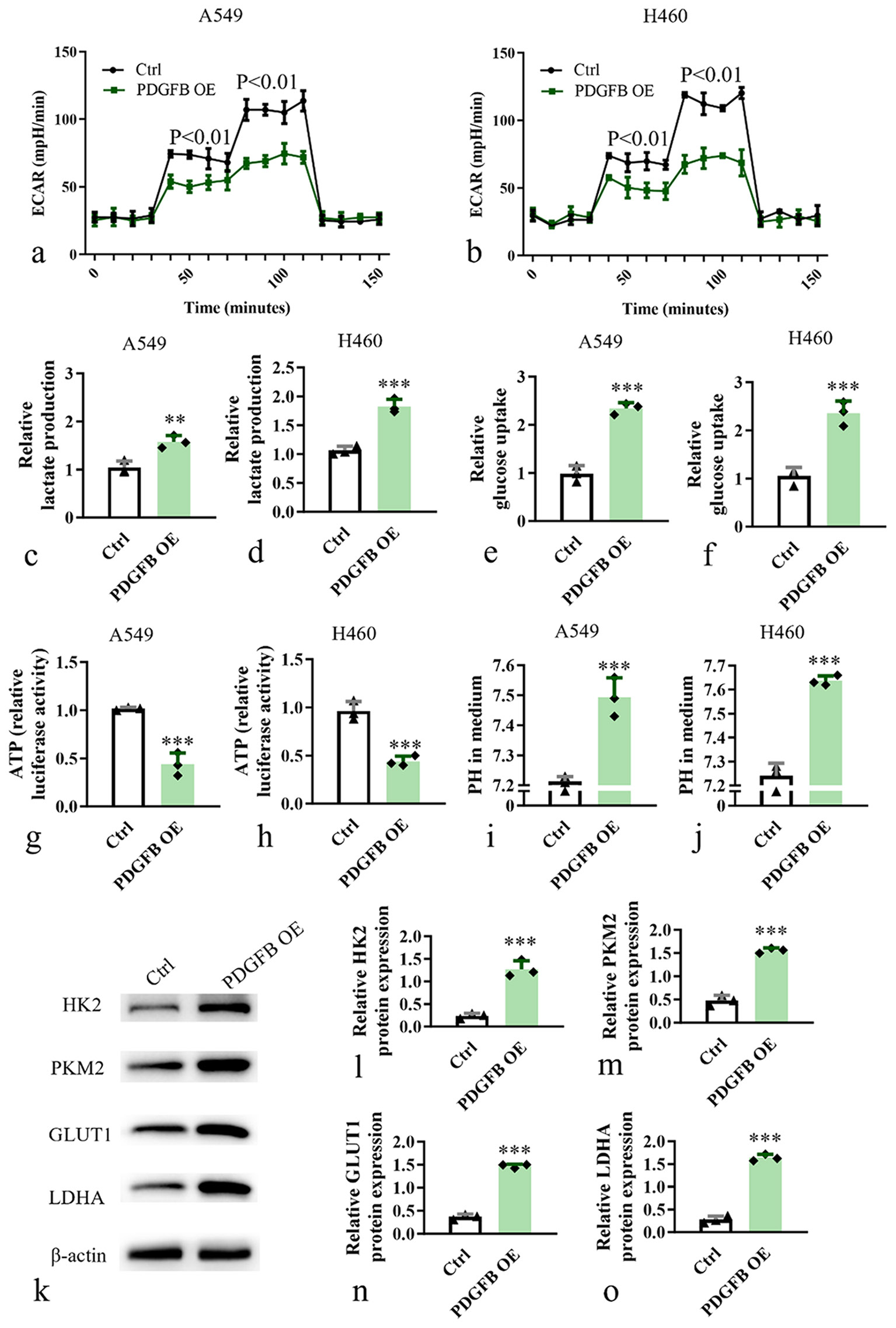
- PDGFB modulates aerobic glycolysis of lung cancer cell. (a) Aerobic glycolysis of A549 cell. (b) Aerobic glycolysis of H460 cell. (c and d) Lactate levels analysis of A549 and H460. (e and f) Glucose uptake analysis of A549 and H460. (g and h) ATP levels analysis of A549 and H460. (i and j) PH value analysis of A549 and H460. (k-o) Western blot analysis of glycolysis-related regulatory protein levels in A549 and H460. n = 3. ✶✶P < 0.01, ✶✶✶P < 0.001. ECAR: Extracellular acidification rate, ATP: Adenosine triphosphate, HK2: Hexokinase 2, PKM2: Pyruvate kinase M2, GLUT1: Facilitative glucose transporter, LDHA: Lactate dehydrogenase, PDGFB: Platelet-derived growth factor subunit B.
PDGFB promoted lung cancer cell growth and metastasis through activating aerobic glycolysis
To test whether the promoting effects of PDGFB on lung cancer cell growth and metastasis were dependent on increased aerobic glycolysis, glucose in the cell culture medium was replaced by galactose, which induced cells to rely on mitochondrial metabolism to generate sufficient ATP for survival. A549 and H460 cells from the PDGFB OE group were treated with galactose to investigate the impact on cell behaviors related to proliferation, migration, and invasion. We employed scratch assays and Transwell assays to examine cell migration [Figure 5a-f], Matrigel invasion assay to assess cell invasion [Figure 5g-i], and clonogenic formation assays to evaluate cell proliferation [Figure 5j-l]. Supplement of the galactose significantly suppressed cell migration (P < 0.001), invasion (P < 0.001), and proliferation (P < 0.001) in the PDGFB OE group. This finding suggests that PDGFB promoted lung cancer cell growth and metastasis through activating aerobic glycolysis. These results provide deeper insights into the molecular mechanisms underlying the influence of PDGFB on lung cancer cell behavior and highlight the potential therapeutic implications of targeting PDGFB for inhibiting lung cancer progression.
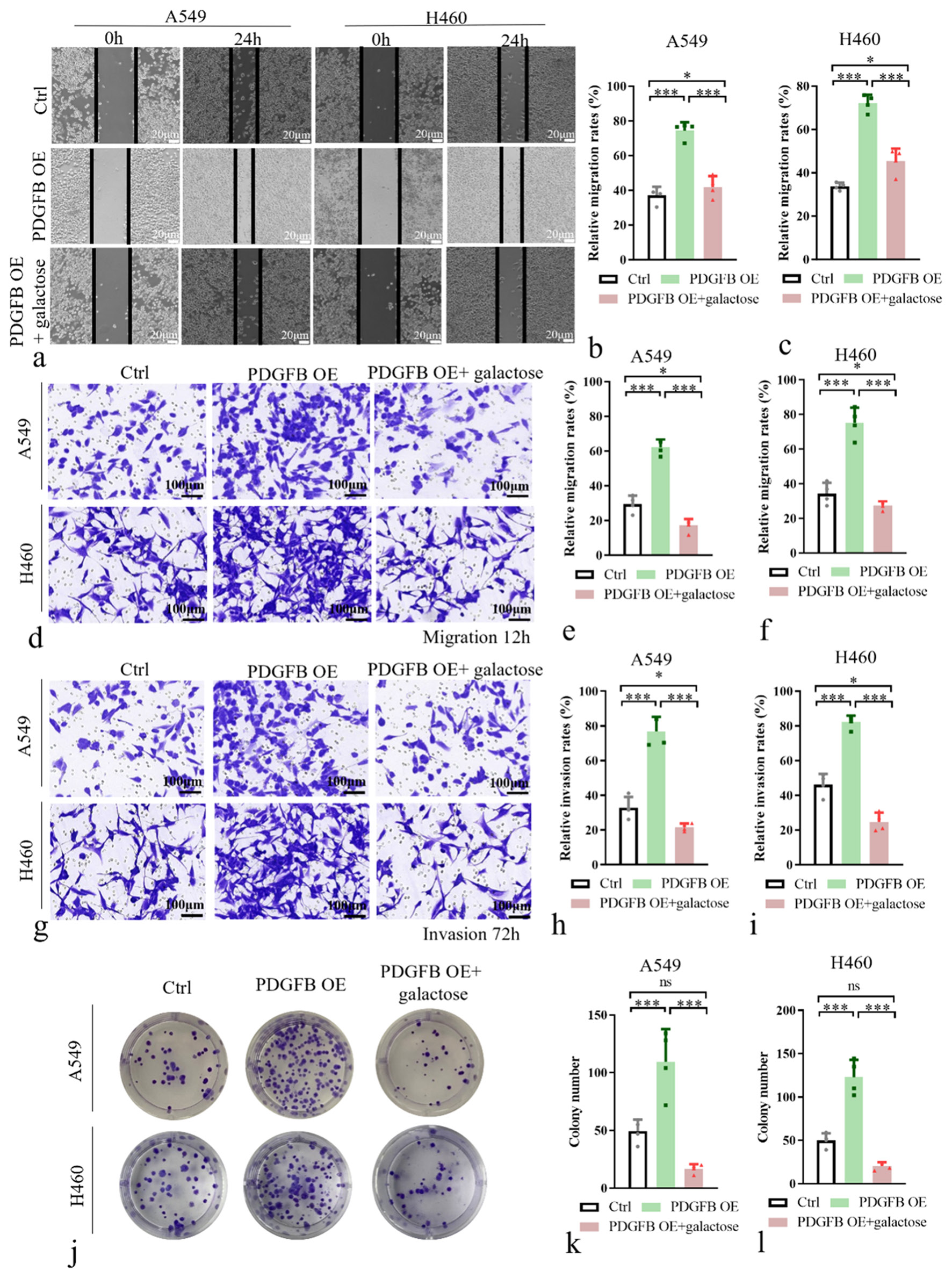
- PDGFB promoted lung cancer cell growth and metastasis through activating aerobic glycolysis. (a-c) Wound-healing cell migration assay with galactose in A549 and H460. (d-f) Transwell chamber assays with galactose in A549 and H460. (g-i) Matrigel invasion assays with galactose in A549 and H460. (j-l) Colony formation assays with galactose in A549 and H460. n = 4. ns: Not significant, ✶P < 0.01, ✶✶✶P < 0.001. PDGFB: Platelet-derived growth factor subunit B.
DISCUSSION
In recent years, extensive research has been conducted to explore the role of overexpressed PDGFB in tumor growth and metastasis.[14,15] PDGFB, as a crucial cytokine, plays a significant role in regulating various biological processes, including cell proliferation,[16] differentiation,[17] and migration.[18] Its aberrant expression and dysregulation have been associated with the development and progression of various cancers, including lung cancer.
Insights from recent studies have shed light on the impact of PDGFB OE on cellular glucose metabolism.[19] Glucose, as a vital energy source, is tightly regulated in cells.[20,21] It has been observed that the overexpression of PDGFB leads to increased glucose uptake capacity and enhanced glycolytic flux, favoring the utilization of glucose for energy production and biomass synthesis.[22]
Our study’s investigation into the impact of PDGFB on lung cancer reveals intricate connections between glucose metabolism and tumor behavior. The significant upregulation of PDGFB in lung cancer cell lines and patient tumor samples suggests a pivotal role for PDGFB in driving the progression of the disease. The findings demonstrate that elevated PDGFB expression is associated with increased cell proliferation, invasion, and migration, highlighting the multifaceted effects of PDGFB on tumor aggressiveness. The correlation between PDGFB levels and changes in glycolysis-related proteins underscores the intricate interplay between PDGFB signaling and metabolic reprogramming in promoting tumor growth.
Understanding the regulatory mechanisms underlying the influence of PDGFB on glucose metabolism in lung cancer cells is of immense value. It offers opportunities for the development of targeted therapeutic strategies to disrupt metabolic adaptations in cancer cells. We assumed that PDGFB can exert its effects on glucose metabolism by regulating key enzymes and transporters involved in glucose uptake and glycolysis. Upregulation of glucose transporters, such as GLUT1, has been observed in lung cancer cells overexpressing PDGFB, facilitating increased glucose entry into the cells. In addition, PDGFB can modulate the expression and activity of enzymes involved in glycolysis, promoting the conversion of glucose into pyruvate and subsequent ATP production. The alterations in glucose metabolism associated with PDGFB OE have significant implications for tumor growth and metastasis. Enhanced glycolysis provides cancer cells with a metabolic advantage, supplying the necessary energy and building blocks for rapid proliferation. Moreover, upregulated glucose metabolism has been linked to the promotion of tumor angiogenesis, invasiveness, and metastatic potential. By selectively targeting PDGFB or crucial enzymes involved in glucose metabolism, it may be possible to suppress tumor growth, inhibit metastasis, and sensitize cancer cells to existing treatment modalities.
Moreover, the study’s ability to attenuate PDGFB-induced effects by targeting aerobic glycolysis provides novel insights into potential therapeutic strategies for lung cancer. By disrupting the metabolic pathways altered by PDGFB, there is a possibility of identifying vulnerabilities that can be exploited for therapeutic benefit. The implications of these findings extend beyond the scope of this study, pointing toward PDGFB as a promising target for precision medicine approaches in lung cancer treatment. The interconnection between PDGFB, glucose metabolism, and tumor behavior offers a new paradigm for developing targeted therapies that address the specific molecular characteristics of individual tumors. Overall, the overexpression of PDGFB in lung cancer cells significantly affects glucose metabolism. Further investigation into the intricate interplay between PDGFB signaling, glucose metabolism, and tumor progression holds promise for advancing our knowledge of cancer biology and therapeutic development.
This study also has limitations. One limitation is the reliance on cell line models, which may not fully recapitulate the complexity of tumor behavior in the human body. While cell line studies offer valuable mechanistic insights, further validation in in-vivo models and clinical settings is necessary to confirm the translational relevance of the findings.
Another limitation is the focus on a specific aspect of PDGFB function and its effects on glucose metabolism. The study primarily investigated the impact of PDGFB OE on cell behavior and metabolic pathways. Future studies could explore additional regulatory mechanisms and downstream signaling pathways associated with PDGFB to provide a comprehensive understanding of its role in lung cancer progression.
In addition, the study’s reliance on inhibition of aerobic glycolysis to mitigate PDGFB-induced effects, while informative, may oversimplify the intricacies of metabolic reprogramming in cancer cells. Further exploration of alternative metabolic pathways and potential compensatory mechanisms activated in response to PDGFB modulation could yield deeper insights into the metabolic vulnerabilities of lung cancer cells.
Despite these limitations, the findings of the study underscore the potential of targeting PDGFB in the context of lung cancer therapy. By unraveling the complex interplay between PDGFB, glucose metabolism, and tumor behavior, this study lays the foundation for future research aimed at developing innovative therapeutic strategies that leverage the vulnerabilities created by metabolic alterations in cancer cells.
SUMMARY
Our study demonstrates that PDGFB plays a crucial role in promoting the growth and metastasis of lung cancer by impacting glucose metabolism. Elevated PDGFB expression significantly enhances lung cancer cell proliferation, invasion, and migration; while, inhibiting aerobic glycolysis effectively counteracts the promoting effects of PDGFB on these cellular processes. These findings emphasize the potential of targeting PDGFB as a therapeutic strategy in lung cancer treatment.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the present study were available from the corresponding author on reasonable request.
AUTHOR CONTRIBUTIONS
KF and GFQ: Designed the research study; XPC, GFQ, and KF: Performed the research; GFQ: Analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
ABBREVIATIONS
AKT: Protein kinase B
ATCC: American Type Culture Collection
ATP: Adenosine triphosphate
DMEM: Dulbecco’s modified eagle medium
ECAR: Extracellular acidification rate assay
EdU: 5-ethynyl-2’-deoxyuridine
GLUT1: Glucose transporter 1
HK2: Hexokinase 2
LDHA: Lactate dehydrogenase A
mTOR: Mammalian target of rapamycin
PDGFB: Platelet-derived growth factor subunit B
PDH: Pyruvate dehydrogenase
PDK1: Pyruvate dehydrogenase kinase 1
PI3K: Phosphoinositide 3-kinase
PKM2: Pyruvate kinase M2
qRT-PCR: Real-time quantitative polymerase chain reaction
STR: Short-tandem repeat analysis
ACKNOWLEDGMENT
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Medical Ethics Committee of Shandong Public Health Clinical Center, Shandong University (approval No. KYLL-20230617) and was performed in accordance with the principles of the Declaration of Helsinki. All eligible participants signed an informed consent form.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: Not applicable.
References
- Lung cancer 2020: Epidemiology, etiology, and prevention. Clin Chest Med. 2020;41:1-24.
- [CrossRef] [PubMed] [Google Scholar]
- Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877-919.
- [CrossRef] [PubMed] [Google Scholar]
- LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol. 2019;12:91.
- [CrossRef] [PubMed] [Google Scholar]
- Tumor aerobic glycolysis confers immune evasion through modulating sensitivity to T cell-mediated bystander killing via TNF-α. Cell Metab. 2023;35:1580-96.e9.
- [CrossRef] [PubMed] [Google Scholar]
- Platelet-specific PDGFB ablation impairs tumor vessel integrity and promotes metastasis. Cancer Res. 2020;80:3345-58.
- [CrossRef] [PubMed] [Google Scholar]
- Diverse roles of tumor-stromal PDGFBto-PDGFRβ signaling in breast cancer growth and metastasis. Adv Cancer Res. 2022;154:93-140.
- [CrossRef] [PubMed] [Google Scholar]
- The PDGF family is associated with activated tumor stroma and poor prognosis in ovarian cancer. Dis Markers. 2022;2022:5940049.
- [CrossRef] [PubMed] [Google Scholar]
- Cancer cell-derived PDGFB stimulates mTORC1 activation in renal carcinoma. Int J Mol Sci. 2023;24:6447.
- [CrossRef] [PubMed] [Google Scholar]
- Milestones in tumor vascularization and its therapeutic targeting. Nat Cancer. 2024;5:827-43.
- [CrossRef] [PubMed] [Google Scholar]
- Proteasome inhibitor bortezomib prevents proliferation and migration of pulmonary arterial smooth muscle cells. Kaohsiung J Med Sci. 2024;40:542-52.
- [CrossRef] [PubMed] [Google Scholar]
- PDGFRβ regulates adipose tissue expansion and glucose metabolism via vascular remodeling in diet-induced obesity. Diabetes. 2017;66:1008-21.
- [CrossRef] [PubMed] [Google Scholar]
- PDGFBB/PDGFRβ promotes epithelial-mesenchymal transition by affecting PI3K/AKT/mTOR-driven aerobic glycolysis in Wilms' tumor G401 cells. Cell Biol Int. 2022;46:907-21.
- [CrossRef] [PubMed] [Google Scholar]
- Declaration of Helsinki: Ethical principles for medical research involving human participants. JAMA. 2025;333(1):71-74.
- [CrossRef] [PubMed] [Google Scholar]
- Tumor-mesothelium HOXA11-PDGF BB/TGF β1-miR-181a-5p-Egr1 feedforward amplifier circuity propels mesothelial fibrosis and peritoneal metastasis of gastric cancer. Oncogene. 2024;43:171-88.
- [CrossRef] [PubMed] [Google Scholar]
- Activated platelets facilitate hematogenous metastasis of breast cancer by modulating the PDGFR-β/COX-2 axis. iScience. 2023;26:107704.
- [CrossRef] [PubMed] [Google Scholar]
- Preosteoclast plays a pathogenic role in syndesmophyte formation of ankylosing spondylitis through the secreted PDGFB-GRB2/ERK/RUNX2 pathway. Arthritis Res Ther. 2023;25:194.
- [CrossRef] [PubMed] [Google Scholar]
- Bone-derived PDGF-BB drives brain vascular calcification in male mice. J Clin Invest. 2023;133:e168447.
- [CrossRef] [PubMed] [Google Scholar]
- Endothelial Twist1-PDGFB signaling mediates hypoxia-induced proliferation and migration of αSMA-positive cells. Sci Rep. 2020;10:7563.
- [CrossRef] [PubMed] [Google Scholar]
- MTMR7 suppresses the phenotypic switching of vascular smooth muscle cell and vascular intimal hyperplasia after injury via regulating p62/mTORC1-mediated glucose metabolism. Atherosclerosis. 2024;390:117470.
- [CrossRef] [PubMed] [Google Scholar]
- Cell-in-cell promotes lung cancer malignancy by enhancing glucose metabolism through mitochondria transfer. Exp Cell Res. 2023;429:113665.
- [CrossRef] [PubMed] [Google Scholar]
- Adipocytes reprogram cancer cell metabolism by diverting glucose towards glycerol-3-phosphate thereby promoting metastasis. Nat Metab. 2023;5:1563-77.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatocyte CYR61 polarizes profibrotic macrophages to orchestrate NASH fibrosis. Sci Transl Med. 2023;15:eade3157.
- [CrossRef] [PubMed] [Google Scholar]








