Translate this page into:
Chloride intracellular channel 6 inhibits hepatocellular carcinoma progression by modulating immune cell balance and promoting tumor cell apoptosis

*Corresponding author: Xueyang Chen, Department of Pathology, Central Hospital Affiliated to Shandong First Medical University, Jinan, China. xychen1995@126.com
-
Received: ,
Accepted: ,
How to cite this article: Zhou H, Xi Y, Chen X. Chloride intracellular channel 6 inhibits hepatocellular carcinoma progression by modulating immune cell balance and promoting tumor cell apoptosis. CytoJournal. 2025;22:20. doi: 10.25259/Cytojournal_183_2024
Abstract
Objective
Chloride intracellular channel 6 (CLIC6) is essential for the development of cancer, and it is widely studied for the treatment of various cancers. This study aimed to explore the potential mechanisms of CLIC6 in the treatment of hepatocellular carcinoma (HCC).
Material and Methods
Initially, a subcutaneous xenograft model of HCC was established. The model groups were treated with varying levels of CLIC6 recombinant protein. After 21 days, tumor and liver tissues were harvested. Tumor size and weight were measured, and hematoxylin-eosin staining was used to assess histopathological changes in the tumor tissues. Terminal deoxynucleotidyl transferase-mediated 2’-deoxyuridine 5’-triphosphate nick-end labeling staining was employed to evaluate apoptosis in tumor tissue cells. Quantitative real-time polymerase chain reaction and Western blot were utilized to analyze cytokine messenger ribonucleic acid ( mRNA) levels in the liver or tumor tissues, and immunohistochemistry was conducted to assess cytokine expression.
Results
CLIC6 significantly inhibits tumor proliferation and enhances apoptosis in tumor tissue cells. CLIC6 markedly reduces the mRNA levels of interleukin (IL)-6, IL-1β, interferon-γ, tumor necrosis factor-α, and IL-17A in liver tissue when increasing transforming growth factor-β and IL-4 mRNA levels. CLIC6 potentially modulates Th cell balance by regulating forkhead box protein P3, GATA-binding protein 3, T-box expressed in T cell, and retinoic acid receptor-related orphan receptor γt (ROR-γt) expression, thereby restraining HCC progression in mice. Moreover, CLIC6 mitigates hepatic oxidative damage via the Janus tyrosine kinase 1/signal transducer and activator of the transcription pathway, attenuates c-Jun N-terminal kinase (JNK) phosphorylation, and modulates apoptosis-related proteins, effectively hindering HCC development.
Conclusion
CLIC6 demonstrates potent antitumor effects in HCC through inhibition of proliferation, promotion of apoptosis, modulation of cytokine levels, regulation of immune cell balance, and attenuation of oxidative stress pathways.
Keywords
Apoptosis
Chloride intracellular channel 6
Hepatocellular carcinoma
INTRODUCTION
The high incidence and mortality rates of liver cancer, primarily hepatocellular carcinoma (HCC), pose a remarkable global health burden, particularly in areas where viral hepatitis and cirrhosis are prevalent chronic liver disorders.[1,2] The prognosis for patients with advanced HCC remains poor, even with advancements in early identification and treatment approaches such as liver transplantation, locoregional treatments, and surgical resection.[3,4] Novel therapeutic strategies that could successfully target tumor progression and enhance patient outcomes are desperately needed.
Chloride intracellular channel 6 (CLIC6) is considered a potentially significant regulatory factor in cancer biology.[5-7] Recent studies have shown that CLIC6 is involved in regulating ion transport, cell signaling, and modulating the tumor microenvironment among various cellular processes.[8] Numerous reports have shown that the JNK signaling pathway regulates liver cancer.[9,10] In addition, research has indicated that CLIC6 is downregulated in HCC.[11] However, its specific role in HCC remains incompletely understood.
This study aimed to comprehensively explore the multifaceted mechanisms by which CLIC6 influences the immune landscape and apoptotic pathways within the HCC microenvironment. Understanding these mechanisms is crucial for optimizing CLIC6’s therapeutic potential in clinical settings, potentially offering new avenues for combination therapies and personalized treatment approaches in managing advanced HCC. CLIC6 has the potential to be a game-changing agent in the battle against liver cancer by bridging the gap between basic scientific findings and clinical applications. This potential could enhance patient outcomes and change how this difficult illness is treated.
MATERIAL AND METHODS
Establishment of liver cancer animal model
Thirty male BALB/c nude mice, aged 6–8 weeks and weighing 25 ± 2 g, were purchased from the Institute of Materia Medica, Chinese Academy of Sciences. This study was approved by the Experimental Animal Welfare Ethics Committee of the Association. In accordance with the random number table method, the nude mice were divided into control group, model group, model + low CLIC6 (10 nM/day, ab106875, Abcam, Cambridge, MA, USA) group, model + medium CLIC6 (20 nM/day) group, and model + high CLIC6 (50 nM/day) group.[12] Pentobarbital sodium (40 mg/kg, 21642-83-1, Shandong Xiya Chemical Industry Co., Ltd., Shandong, China) was injected intraperitoneally to anesthetize the nude mice. Using a 1 mL sterile syringe, an appropriate amount of HepG2 cell (iCell-h092, iCELL, Shanghai, China) suspension (approximately 100 μL, containing 2 × 106 cells) was slowly injected subcutaneously into the back of the nude mice to establish a subcutaneous xenograft liver cancer model. Caution was taken to ensure that the cell suspension was evenly distributed without forming bubbles. After the injection, the nude mice were returned to sterile cages, and their health status was monitored. Tumor growth was regularly observed. Different doses of CLIC6 were administered to the model + low CLIC6 (10 nM/day) group, model + medium CLIC6 (20 nM/day) group, and model + high CLIC6 (50 nM/day) group via tail vein injection on the basis of the model group. A daily injection of an equivalent volume of saline was administered to the control group. The nude mice were sacrificed using an intraperitoneal dose of pentobarbital sodium (110 mg/kg) following a 21-day experiment. Tumor and liver tissues were collected for further analysis.
ELISA assay
The serum levels of mice were analyzed with NO (S0021S), malondialdehyde (MDA, S0131S), and super oxide dismutase (SOD, S0101S) ELISA kits. Kits purchased from Beyotime Biotechnology (Shanghai, China). Perform experiments according to kit instructions. The analysis was carried out by enzyme-labeled apparatus (Multiskan SkyHigh, Thermo Fisher Scientific, Waltham, MA, USA).
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining
Tissue samples were embedded in paraffin. The slides were deparaffinized by immersing them in xylene (2 times, 5 min each, 534056, Merck, Darmstadt, Germany) and then rehydrated through a series of ethanol solutions: 100%, 95%, 85%, and 70% (2 min each). Finally, they were rinsed with distilled water. The sections were incubated with proteinase K solution (20 μg/mL in phosphate buffer saline (PBS), 10412ES, Yeasen, Shanghai, China) for 15–30 min at room temperature and then rinsed with PBS. The sections were treated with 0.1% Triton X-100 (P0096, Beyotime Biotechnology, Shanghai, China) in PBS for 5–10 min at room temperature and then rinsed with PBS. A TUNEL reaction (C1091, Beyotime Biotechnology, Shanghai, China) mixture was prepared in accordance with the manufacturer’s instructions, usually containing terminal deoxynucleotidyl transferase (TdT) enzyme and biotinylated nucleotide mix. This mixture was applied to the sections and incubated in a humidified chamber at 37°C for 1 h. The sections were rinsed with PBS to stop the reaction, incubated with streptavidin- horseradish peroxidase ( HRP) conjugate for 30 min at room temperature, and rinsed with PBS. The sections were mounted with an appropriate mounting medium and covered with coverslips. They were observed under a light microscope (CX53, Olympus, Tokyo, Japan).
Immunohistochemistry
First, tissue samples were embedded in paraffin, or sections were prepared. Then, the slides were immersed in xylene to deparaffinize (2 times, 5 min each), rehydrated through a series of ethanol solutions (100%, 95%, 85%, and 70%, 2 min each), and rinsed with distilled water. Next, the sections were heated in citrate buffer (pH 6.0, C999, Sigma, St. Louis, MO, USA) or ethylene diamine tetraacetic acid (EDTA) buffer (pH 9.0, 4008M, Sigma, St. Louis, MO, USA) for 10–20 min for antigen retrieval, cooled to room temperature, and rinsed with PBS. The sections were rinsed with PBS again after 10 min of immersion in 3% H2O2 at room temperature to inhibit endogenous peroxidase activity. Then, the sections were incubated in normal serum (such as 10% goat serum or horse serum) for 30 min to block nonspecific binding sites. Subsequently, the sections were incubated with specific primary antibodies for phosph-JNK (1:500, ab307802), T-box expressed in T cell (T-bet) (1:500, ab307193), or cleaved caspase-3 (1:500, ab32042), typically overnight at 4°C or for 1–2 h at room temperature, and then rinsed with PBS. Next, the sections were incubated with secondary antibodies (1:1000, ab6721) for 30 min at room temperature and rinsed with PBS. All antibodies were obtained from Abcam (Cambridge, MA, USA). A 3,3’-diaminobenzidine ( DAB) substrate kit (ml095670, Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China) was used for color development until a brown color appeared (usually 5–10 min) and rinsed with tap water to stop the reaction. The sections were counterstained with hematoxylin (C0107, Beyotime Biotechnology, Shanghai, China) for 1–2 min to stain the nuclei blue and rinsed with tap water. Finally, the sections were mounted with an appropriate mounting medium and covered with coverslips. They were observed under a light microscope (CX53, Olympus, Tokyo, Japan). The images were analyzed by ImageJ (version 1.3.4, National Institutes of Health, Bethesda, MD, USA), and the results were calculated in accordance with integrated optical density (IOD)/area.
RNA extraction and quantitative real-time polymerase chain reaction (PCR)
Total ribonucleic acid ( RNA) was extracted from tissue samples using a TRIzol universal reagent (DP424, TIANGEN, Beijing, China). Subsequently, the RNA was transcribed into complementary DNA (cDNA) using reverse transcriptase (KR103, TIANGEN, Beijing, China), with the addition of specific primers to enhance cDNA synthesis efficiency. Specific primer pairs were designed, with one serving as the forward primer and the other as the reverse primer for subsequent PCR amplification of the cDNA. The prepared PCR reaction mixture included the cDNA template, primers, DNA polymerase, and other necessary reagents and involved cycles of denaturation, annealing, and extension. Real-time monitoring of fluorescence signals during PCR reflected proportional amplification of PCR products. Data analysis involved quantifying target gene expression levels using standard curves or the 2−ΔΔCt method. Finally, the obtained data were compared and interpreted relative to the reference gene (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) to assess gene expression differences between samples. The primer sequences employed are detailed in Supplementary Table 1.
Western blot
First, the protein in the sample was extracted with radioimmunoprecipitation assay ( RIPA, P0013B, Beyotime Biotechnology, Shanghai, China), and its concentration was measured with a bicinchoninic acid assay kit (P0011, Beyotime Biotechnology, Shanghai, China) to ensure quality. Next, the proteins were separated by size through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis and transferred onto polyvinylidene fluoride (IPVH00010, Millipore Corporation, MA, USA) or nitrocellulose membrane. The membrane was then incubated with specific primary antibodies phosph- signal transducer and activator of transcription (p-STAT) 3 (1:1000, ab267373), signal transducer and activator of transcription (STAT) 3 (1:1000, ab 68153), p-p38 (1:1000, ab195049), p38 (1:1000, ab170099), phospho-janus kinase (p-JAK) 1 (1:1000, ab138005), Janus kinase (JAK) 1 (1:1000, ab133666), CLIC6 (1:1000, PA5-101519, Thermo Fisher Scientific, Waltham, MA, USA), and GAPDH (1:1000, ab9485) washed with buffer after binding the target protein. All primary antibodies were obtained from Abcam (Cambridge, MA, USA). Subsequently, a secondary antibody labeled with an enzyme or fluorescent substance was applied, followed by another washing with buffer to remove unbound secondary antibodies (1:2000 dilution; cat nos. ZB-2305, ZB-2301, ZSGB-BIO, Beijing, China). Finally, gel imaging systems were used to capture the staining signals for analysis. Typically, results are compared to a reference protein (GAPDH) to ensure data accuracy and reliability. An enhanced chemiluminescence kit (BL520b, Biosharp Life Science, Hefei, Anhui, China) was used to develop the protein, and ImageJ (version 1.3.4, National Institutes of Health, Bethesda, MD, USA) was used to analyze the gray values of protein bands.
Cell culture
HepG2 cells (iCell-h092) were purchased from iCell (Shanghai, China). Dulbecco’s modified eagle medium (DMEM) was supplemented with 10% fetal bovine serum (C0226, Beyotime Biotechnology, Shanghai, China) and 1% penicillin-streptomycin solution (C0222, Beyotime Biotechnology, Shanghai, China). The frozen HepG2 cells were thawed, transferred to a centrifuge tube containing the medium, and centrifuged. The supernatant was discarded, and the cells were resuspended and cultured at 37°C with 5% carbon dioxide. When the cell confluence reached 80–90%, the cells were passaged, washed with PBS, and incubated with trypsin-EDTA solution. The cells were added with medium to neutralize the trypsin, centrifuged, resuspended, and passaged at a 1:3 or 1:5 ratio. For cell cryopreservation, the cells were resuspended in a freezing solution containing 90% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO), aliquoted into cryovials, placed in a −80°C freezer overnight, and transferred to a liquid nitrogen tank for long-term storage. The cells used in this study underwent short tandem repeat (STR) authentication and were tested negative for mycoplasma.
5-Ethynyl-2'-deoxyuridine (EdU) staining of HepG2 cells
First, HepG2 cells were cultured in a 96-well plate until the cell confluence reached 70–80%. Subsequently, EdU solution (10 μM, E-CK-A377, Elabscience, Wuhan, China) was added to each well in accordance with the experimental design, and the wells were incubated for 1–2 h to promote cellular uptake. After the cells were fixed with 4% paraformaldehyde, they were washed with PBS. Next, a 0.5% Triton X-100 solution was used to permeabilize the cells, and the cells were given another PBS wash. EdU staining reaction (C0085S, Beyotime Biotechnology, Shanghai, China) was implemented using a click chemistry reaction kit, and the cells were incubated at room temperature in the dark for 30 min. After being stained, the cells were washed with PBS and optionally stained with 4',6-diamidino-2'-phenylindole (DAPI) to label the cell nuclei. Finally, the images of EdU-positive cells were observed and captured using a fluorescence microscope (BX53, Olympus, Tokyo, Japan).
TUNEL staining of HepG2 cells
First, HepG2 cells were fixed with 4% paraformaldehyde to preserve their morphology. Subsequently, the cells were permeabilized using a permeabilization agent, such as 0.1% Triton X-100, to allow the TUNEL reaction mixture to enter the cells. Next, a reaction mixture containing TdT enzyme and fluorescently labeled dUTP was added to the samples, typically incubating at room temperature for 30 min. The reaction was terminated by adding a stop reaction solution. Finally, the cell nuclei were optionally stained with DAPI, and the results were observed and analyzed using a fluorescence microscope (BX53, Olympus, Tokyo, Japan).
Statistical analyses
The experimental data were statistically analyzed to assess significant differences between conditions and ensure data reliability. Each experiment was replicated at least 3 times, and the results are presented as the mean ± standard error of the mean. Data processing and visualization were performed using Statistical Package for the Social Sciences software (version 17.0, IBM Corp., Armonk, NY, USA). Analysis of variance was used for intergroup comparisons, with Tukey’s post hoc test or Bonferroni correction applied on the basis of experimental design. P < 0.05 was considered statistically significant.
RESULTS
CLIC6 inhibited the growth and development of liver cancer tumor tissues
A subcutaneous tumor-bearing mice model was established, and tumors were obtained, as shown in Figure 1. The results in Figure 1a-c indicate that the tumor size, volume, and weight of the model group were significantly larger than those of the high-, medium-, and low-dose CLIC6 treatment groups (P < 0.001). The tumor size, volume, and weight gradually decreased with increasing doses of CLIC6, with the high-dose CLIC6 treatment group showing the most pronounced therapeutic effect (P < 0.001). The hematoxylineosin staining results [Figure 1d] showed that compared with the model group, the tumor tissues of the CLIC6 treatment groups with different doses exhibited varying degrees of damage, increased gap, and disorganized cell arrangement, with the high-dose CLIC6 treatment group demonstrating the most significant effect (arrows are shown in Figure 1).
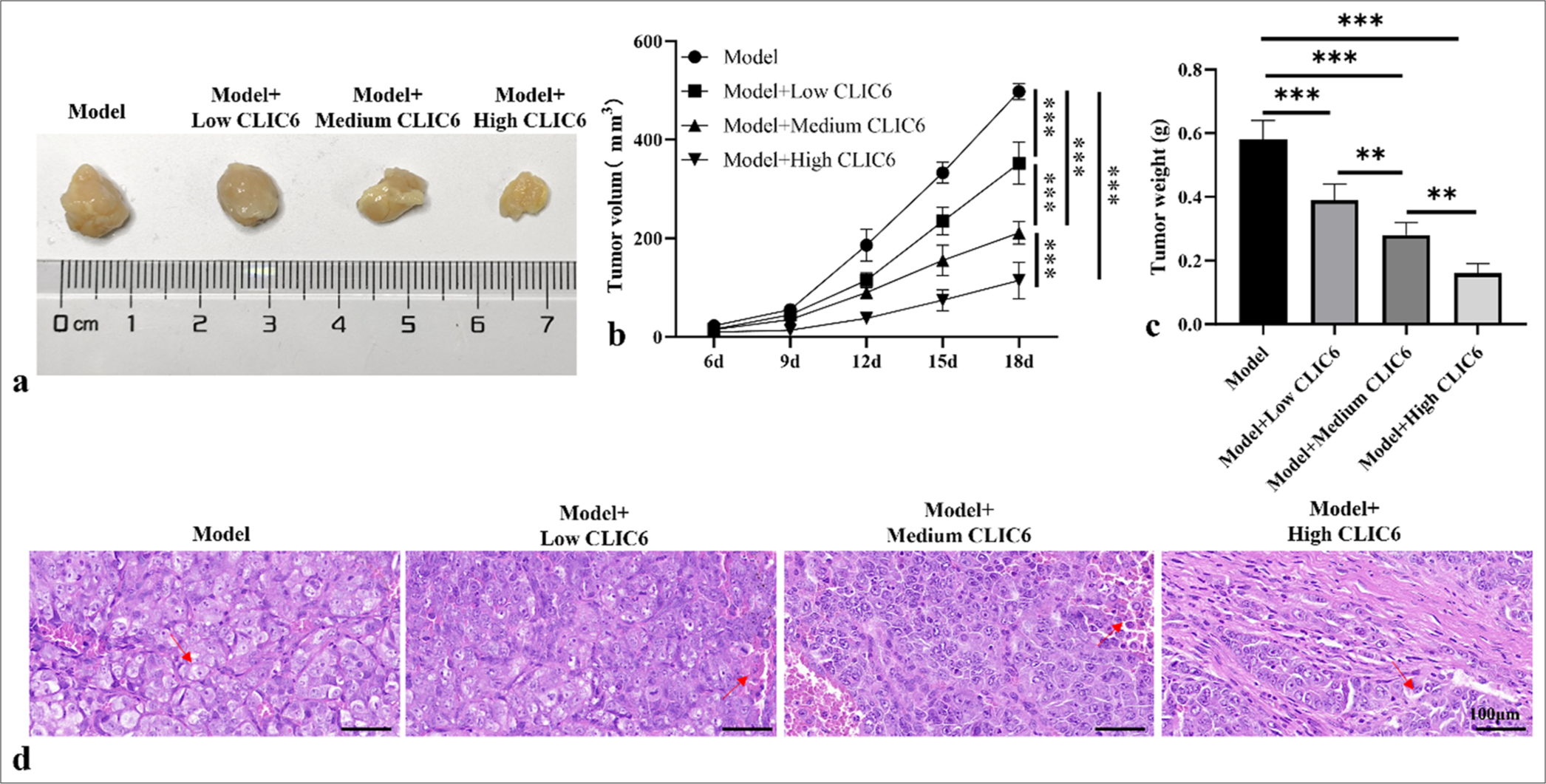
- CLIC6 suppressed the growth and development of liver cancer tumor tissues. (a) Representative images of liver cancer tumors after treatment with different doses of CLIC6. (b) Tumor volume. (c) Tumor weight. (d) Tumor tissues examined using HE staining following varying dosages of CLIC6 intervention, objectve: 200×. (n = 3). Scale bar: 100 μm. n = 6 (✶✶P < 0.01 and ✶✶✶P < 0.001). CLIC6: Chloride intracellular channel 6, HE: Hematoxylin–eosin.
CLIC6 promoted the apoptosis of HCC cells
In Figures 2a and b, the expression of cleaved caspase-3 protein increased significantly at different doses of CLIC6 (P < 0.001). CLIC6 treatment markedly increased the Bax mRNA level and lowered the Bcl-2 mRNA level (P < 0.01 and P < 0.001; [Figure 2c]). The high dose of CLIC6 (20 nM) showed a more significant therapeutic effect than the low and medium doses. The TUNEL staining results [Figure 2d and e] showed that the apoptosis level of tumor tissue treated with high, medium, and low doses of CLIC6 significantly increased (P < 0.05 and P < 0.001, respectively). Among them, the CLIC6 (20 nM) treatment group showed the highest level of apoptosis (P < 0.001).
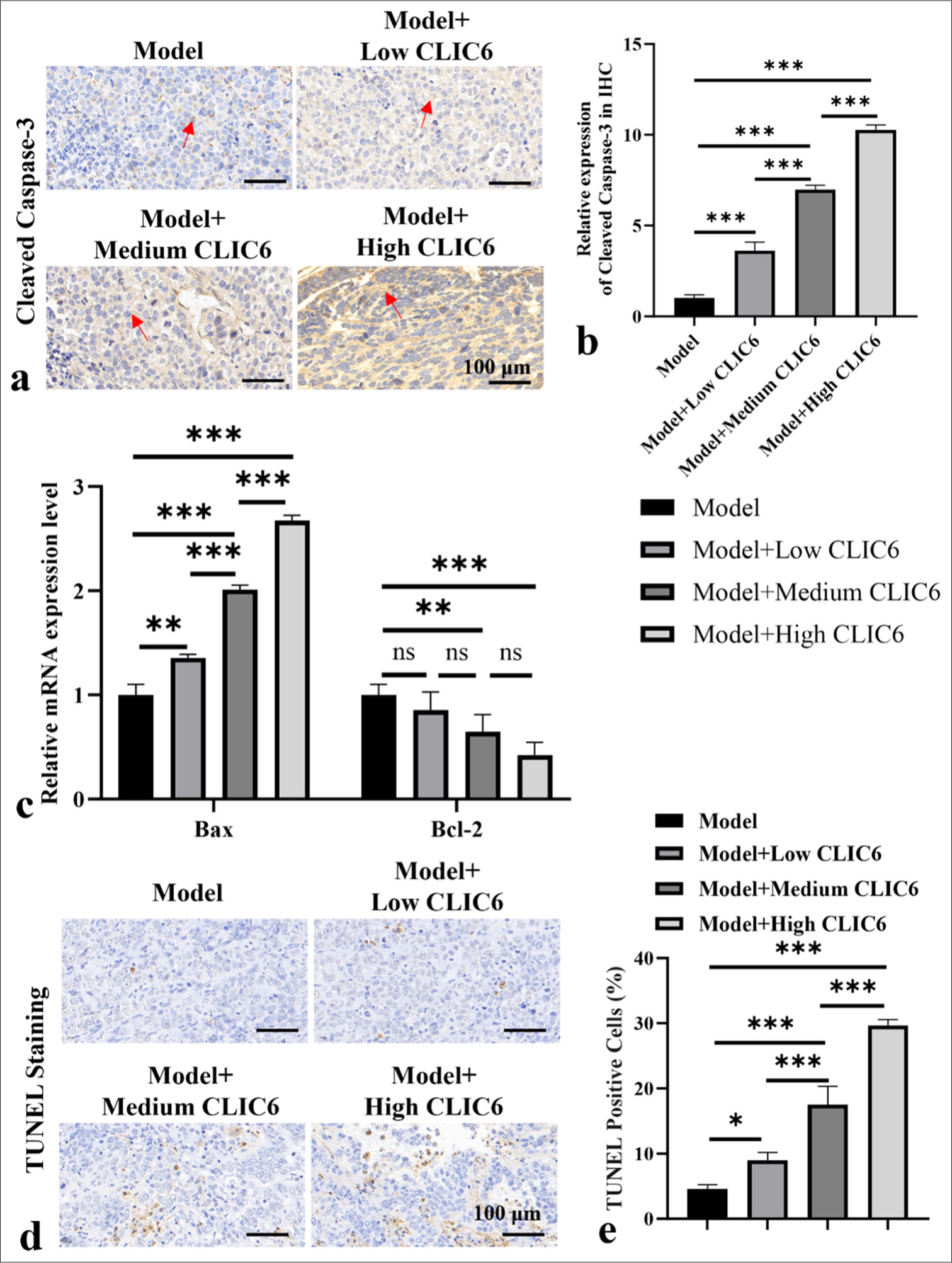
- CLIC6 enhanced apoptosis of hepatocellular carcinoma cells. (a and b) Immunohistochemistry staining of cleaved caspase-3. Scale bar: 100 μm. (c) mRNA levels of Bcl-2 and Bax in tumor tissues. (d and e) TUNEL staining, objectve: 200×, detection of apoptosis level in mice liver cancer tumor tissues. Scale bar: 100 μm. n = 3 (ns: No significant difference; ✶P < 0.05, ✶✶P < 0.01, and ✶✶✶P < 0.001). CLIC6: Chloride intracellular channel 6, TUNEL: Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling, Bcl-2: B-cell lymphoma-2, BAX: Bcl-2-associated X, mRNA: messenger RibonucleicAcid.
CLIC6 is involved in regulating inflammatory response and immune cell equilibrium
As shown in Figure 3a-c, the CLIC6 expression was measured at the molecular and mRNA levels, and it was positively correlated with the dose (P < 0.05 and P < 0.001, respectively). In Figure 3d-j, the levels of Interleukin (IL)-1β, IL-6, tumor necrosis factor-α (TNF-α), interferon (IFN)-γ, and IL-17A in the liver tissue of the model group were higher (P < 0.001). However, the mRNA levels of IL-4 in the 5 nM CLIC6 group decreased (P < 0.05). The mRNA levels of IL-1β, IL-6, TNF-α, IFN-γ, and IL-17A in the 5 nM CLIC6 group decreased (P < 0.05, P < 0.01, and P < 0.001). Meanwhile, these mRNA levels in the medium- and high-dose CLIC6 treatment groups (10 and 20 nM) significantly decreased, whereas the IL-4 and transforming growth factor-β (TGF-β) mRNA levels significantly increased (P < 0.001). In Figure 3k and l, the positive expression of T-bet in the model group was significantly high (P < 0.001). However, the T-bet expression was clearly reduced in all CLIC6 and positive drug treatment groups (P < 0.001). Figure 3m demonstrates that the ROR-γO expression was dramatically downregulated (P < 0.001) in the 20 nM CLIC6 group and significantly upregulated (P < 0.01) in the liver. The expression of ROR-γt was distinctly downregulated in the 20 nM CLIC6 group (P < 0.001). The expression levels of GATA-binding protein 3 (GATA3) and forkhead box protein P3 (Foxp3) in the model group significantly decreased (P < 0.001). The Foxp3 and GATA3 levels significantly increased after 20 nM of CLIC6 treatment (P < 0.001). In addition, CLIC6 can regulate the secretion of immune T cells.
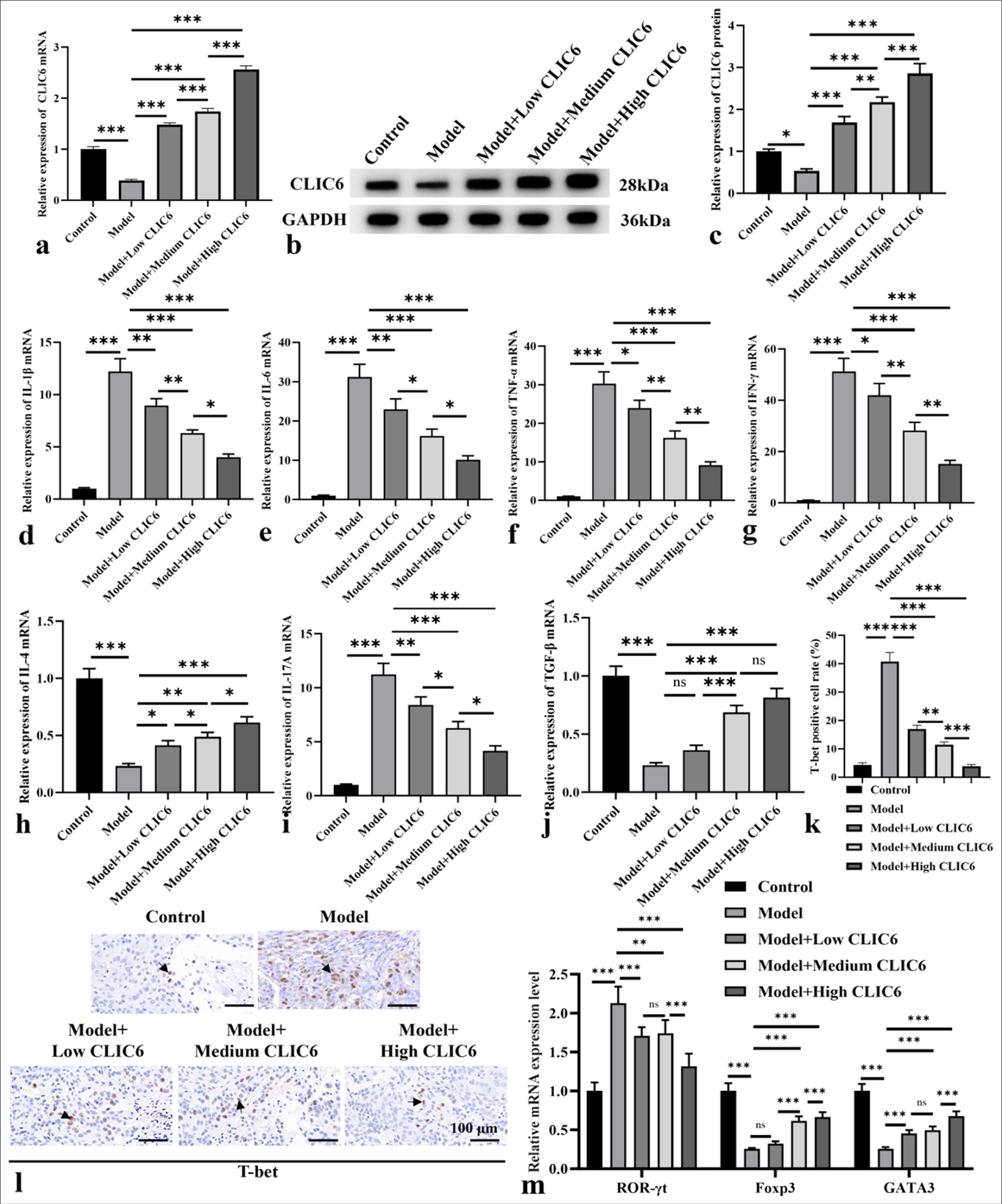
- CLIC6 is involved in inflammatory response and immune cell balance in liver tissue. (a-c) CLIC6 level in liver analyzed by Western blot and qRT-PCR. (d-j) mRNA levels of IL-1β, TNF-α, IFN-γ, IL-6, IL-17A, IL-4, and TGF-β in liver. (k and l) T-bet staining, objectve: 200×, expression in mice liver. Scale bar: 100 μm. (m) Levels of ROR-γt, Foxp3, and GATA3 in liver. n = 3 (ns: No significant difference; ✶P < 0.05, ✶✶P < 0.01, and ✶✶✶P < 0.001). CLIC6: Chloride intracellular channel 6, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, IL: Interleukin, TNF: Tumor necrosis factor, IFN: Interferon, TGF: Transforming growth factor, T-bet: T-box expressed in T cell, ROR-γt: Retinoic acid receptor-related orphan receptor gamma-t, Foxp3: Forkhead box protein P3, GATA3: GATA-binding protein 3, qRT-PCR: Quantitative real-time polymerase chain reaction.
CLIC6 influenced the progression of liver cancer by improving p-JNK-mediated oxidative stress
The liver tissue of the model group demonstrated considerably high p-JNK expression, as shown in Figures 4a and b. Different doses of CLIC6 significantly reduced the positive expression of p-JNK (P < 0.001). In Figures 4c and d, the phosphorylation expression levels of p38 and STAT3 significantly increased (P < 0.001). However, medium and high doses of CLIC6 significantly reduced the p-STAT3 and p-p38 protein levels in the mice liver (P < 0.001). As shown in Figure 4e-g, the serum MDA and NO levels in the model group significantly increased, whereas the SOD level significantly decreased (P < 0.001). The serum MDA and NO levels in the medium- and high-dose CLIC6 groups significantly decreased (P < 0.05), and the SOD level significantly increased (P < 0.01).
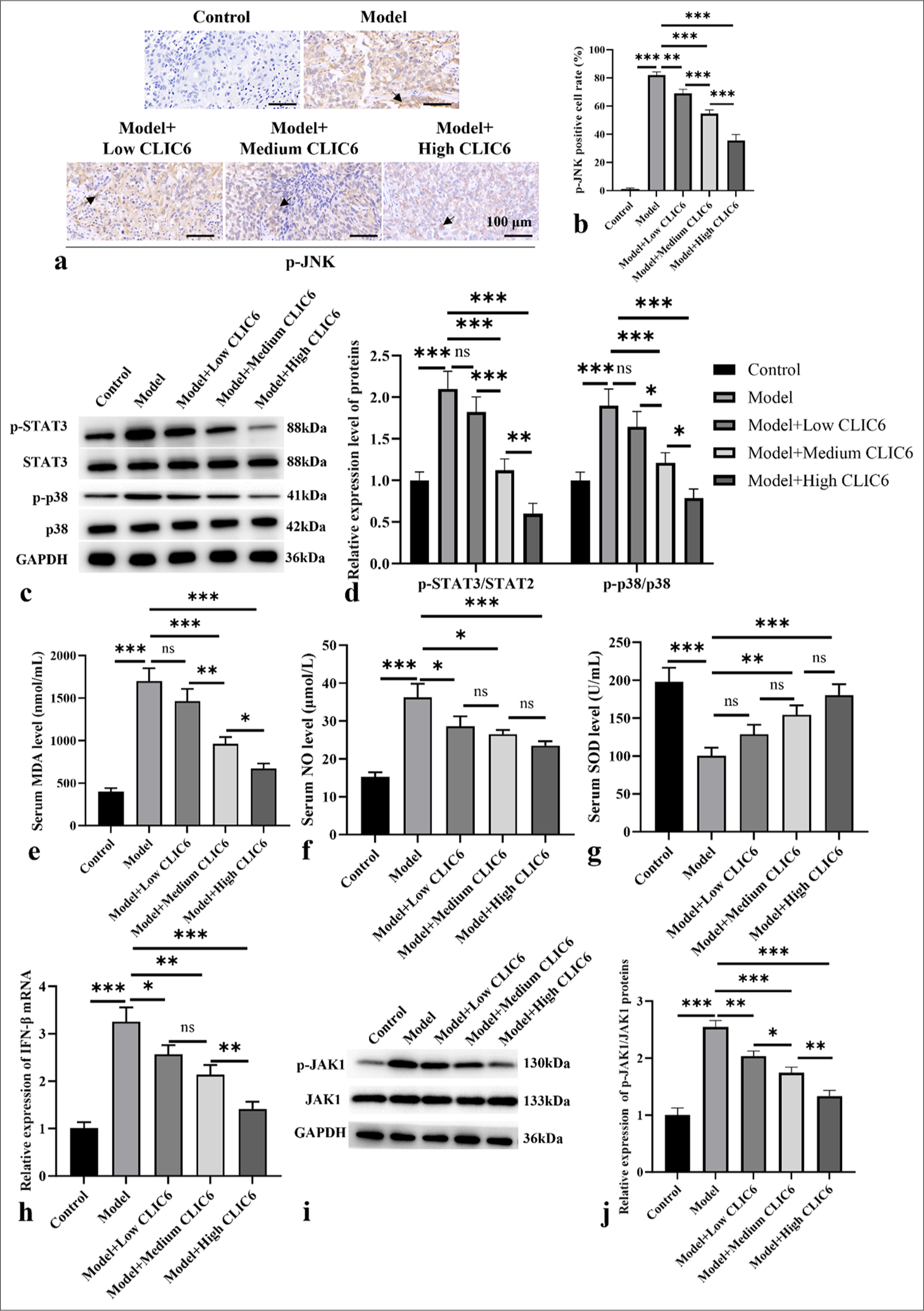
- CLIC6 is involved in p-JNK-mediated oxidant stress. (a and b) Immunohistochemistry staining, objectve: 200×, of p-JNK. Scale bar: 100 μm. (c and d) Expression levels of p-STAT3 and p-p38 proteins in liver. (e-g) Serum levels of MDA (e), NO (f), and SOD(g). (h) IFN-β mRNA levels in liver. (i and j) Protein expression levels of JAK1 and p-JAK1. n = 3 (ns: No significant difference; ✶P < 0.05, ✶✶P < 0.01, and ✶✶✶P < 0.001). CLIC6: Chloride intracellular channel 6, JAK: Janus tyrosine kinase, STAT: Signal transducer and activator of transcription, p38: P38 mitogen-activated protein kinase, IFN: Interferon, p-JNK: phosphor-c-Jun N-terminal kinase, MDA: Malondialdehyde, SOD: Super Oxide Dismutase.
Meanwhile, the expression of IFN-β mRNA in the model group increased (P < 0.001). CLIC6 treatment at different doses significantly reduced the level of IFN-β mRNA (P < 0.05 [Figure 4h]). The p-JAK1 protein expression of the control group was noticeably high (P < 0.001 [Figure 4i and j]). The p-JAK1 protein levels in the CLIC6 groups were reduced (P < 0.01).
CLIC6 inhibited the proliferation of HepG2 cells and promoted apoptosis
Next, the role of CLIC6 in inhibiting liver cancer development was further confirmed at the cellular level in vivo. Figure 5a shows that 10 and 20 nM of CLIC6 significantly inhibited the activity of HepG2 cells in a dose-dependent manner (P < 0.001), with the highest dose of CLIC6 recombinant protein resulting in the lowest cell activity. In Figure 5b-d, with the increase in CLIC6 dose, the CLIC6 level in cells increased significantly (P < 0.05 and P < 0.001). As shown in Figures 5e and f, EdU staining showed a negative correlation between CLIC6 dose and cell proliferation. The TUNEL staining results [Figure 5g and h] indicated that treatment with CLIC6 recombinant protein significantly promoted apoptosis in HepG2 cells, with the high-dose CLIC6 group showing the most pronounced effect (P < 0.001).
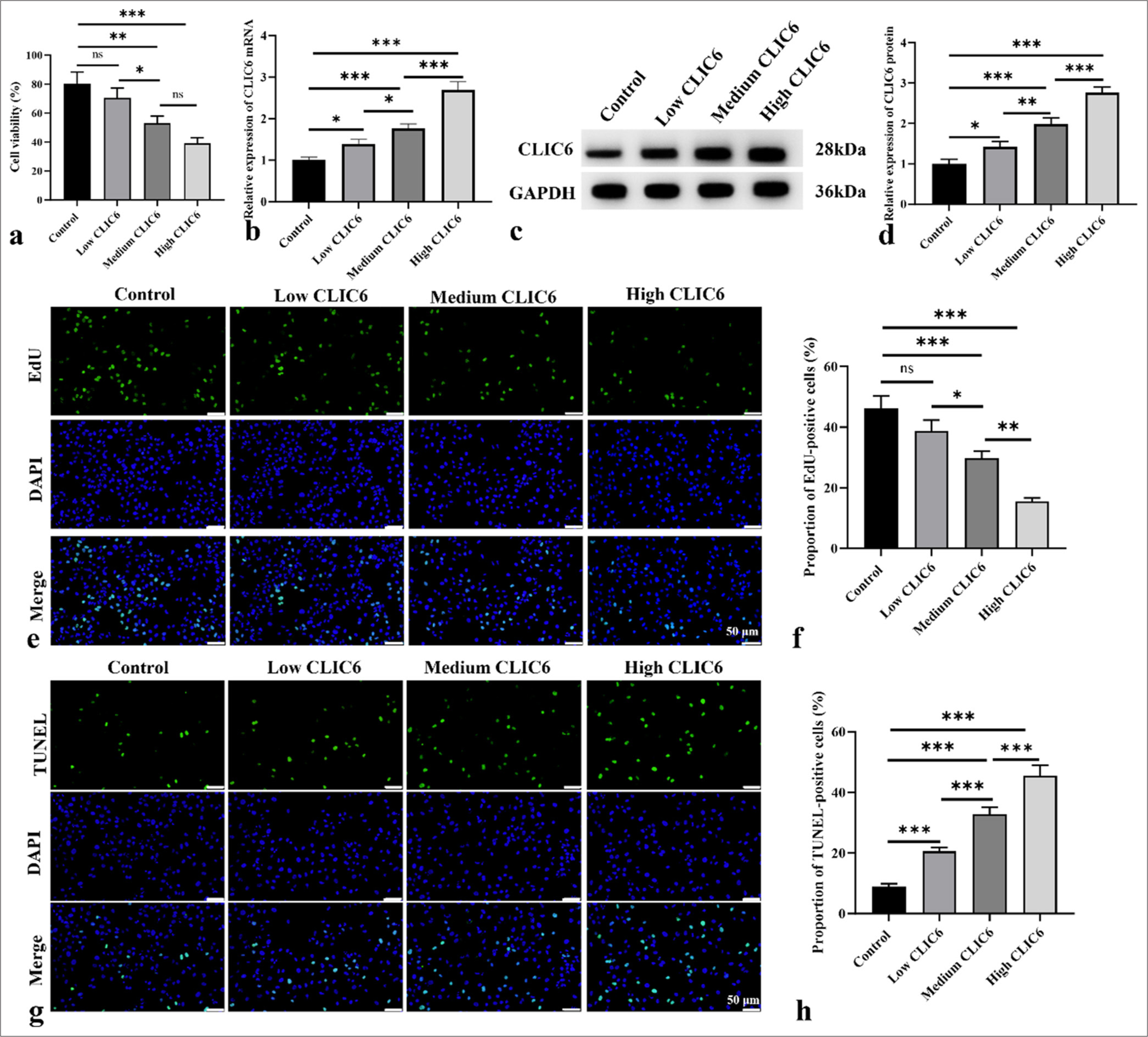
- CLIC6 suppressed proliferation of HepG2 cells and induced apoptosis. (a) Activity of HepG2 cells treated with different doses of CLIC6 recombinant protein. (b-d) Levels of CLIC6 in cells were analyzed by Western Blot and qRT-PCR. (e and f) EdU staining, objectve: 200×, was used to analyze the effects of different doses of CLIC6 recombinant protein on the proliferation capacity of HepG2 cells. Scale bar: 50 μm. (g and h) Cell apoptosis measured using TUNEL staining, objectve: 200×, after treating HepG2 cells with different doses of CLIC6 recombinant protein. Scale bar: 50 μm. n = 3 (ns: No significant difference; ✶P < 0.05, ✶✶P < 0.01, and ✶✶✶P < 0.001). CLIC6: Chloride intracellular channel 6, EdU: 5-ethynyl-2'-deoxyuridine, DAPI: 4',6-diamidino-2'-phenylindole, qRT-PCR: Quantitative real-time polymerase chain reaction, TUNEL: Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
DISCUSSION
This study utilized a subcutaneous tumor-bearing mice model to explore the potential of CLIC6 in the treatment of HCC. The results support the possible involvement of CLIC6 in cancer therapy. Previous studies have indicated that CLIC6 exhibits similar antitumor effects in other cancer types.[13] The results demonstrated that CLIC6 treatment significantly inhibited the growth and metastasis of HCC. The molecular analysis showed that CLIC6 treatment enhanced apoptosis in tumor tissues, as evidenced by increased expression of cleaved caspase-3 protein and upregulation of the Bax/Bcl-2 ratio. These changes indicated that CLIC6 promotes apoptosis of tumor cells, which is crucial for inhibiting tumor progression.
By encouraging the synthesis of anti-inflammatory molecules and controlling the polarization state of immune cells, this work showed that CLIC6 therapy dramatically altered the expression levels of inflammatory cytokines in the tumor microenvironment of HCC. Related research suggests that CLIC6 may inhibit tumor progression by suppressing the expression levels of inflammatory cytokines, such as IL-1β, IL-6, and TNF-α while promoting the generation of immunosuppressive cells such as Treg cells.[7,14] When TGF-β and IL-6 act together on naive CD4+ T cells, they activate the transcription factor retinoic acid receptor-related orphan receptor γt (ROR-γt), promoting the transformation of naive CD4+ T cells into Th17 cells while inhibiting Th 1 and Th 2 cells.[15] Th17 cells are early responding immune cells that defend against various bacteria, acid-fast Mycobacterium tuberculosis, and fungi and deal with pathogens that Th 1 or Th 2 immunity cannot handle.[16] CLIC6 treatment inhibited the expression levels of transcription factors associated with pro-inflammatory Th1 (T-bet) and Th17 (ROR-γt) cells while promoting those of anti-inflammatory Treg cells (Foxp3 and GATA3). These findings suggest that CLIC6 may promote immune cell balance towards a less inflammatory and more regulatory environment, thereby supporting its antitumor effects.
This study also found that CLIC6 affected the phosphorylation levels of STAT3, p38, and JAK1 and was negatively correlated with the dose of CLIC6. The results showed that high doses of CLIC6 could significantly inhibit the phosphorylation of STAT3, p38, and JAK1. In addition, CLIC6 could inhibit the positive expression of p-JNK. The above experimental results showed that CLIC6 may alleviate liver oxidative damage through the JAK1/STAT1 pathway, weaken JNK phosphorylation, promote apoptosis of liver cancer cells, and play a role in alleviating the occurrence of liver cancer. The JAK2/STAT3 pathway is a key factor in the regulation of autophagy, cell death, and cell proliferation. Abnormal JAK2/STAT3 function is involved in almost every stage of cancer development and progression. Many direct and indirect effects of this pathway on the development of malignant tumors have been recognized.[17]
This study also found that CLIC6 treatment restores the levels of antioxidant enzymes and reduces oxidative stress markers (MDA and NO) in serum. Similarly, other studies have indicated that CLIC6 suppresses tumor growth and metastasis by regulating oxidative stress responses.[18] Comparing the mechanisms of CLIC6 in regulating oxidative stress responses across different studies can reveal its comprehensive effects in antitumor therapy.
Comparison with existing literature highlights the novel findings regarding the role of CLIC6 in HCC. CLIC6’s broad therapeutic potential across several malignancies is supported by prior research that found similar antitumor effects in other cancer types.[19] However, the specific mechanisms of CLIC6 in HCC still require further investigation, particularly its interactions with unique immune cells and the signaling pathways specific to liver cancer.
This study comprehensively demonstrated that CLIC6 inhibits the progression of HCC by modulating immune cell balance, promoting tumor cell apoptosis, and regulating oxidative stress and signaling pathways. The findings underscore CLIC6 as a potential therapeutic target for treating HCC, offering new avenues for liver cancer management. Future research should focus on elucidating the precise molecular mechanisms of CLIC6 and conducting clinical trials to validate its therapeutic efficacy in patients with HCC.
SUMMARY
CLIC6 is an antitumor biomarker for HCC. It not only directly prevents liver cancer cells from proliferating and surviving but also alters the tumor microenvironment, encourages tumor cell death, and possesses strong immunomodulatory and antioxidant properties.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study were available from the corresponding author on reasonable request.
ABBREVIATIONS
BAX: BCL2-Associated X
Bcl-2: B-cell lymphoma-2
CLIC6: Chloride intracellular channel 6
Edu: 5-ethynyl-2’-deoxyuridine
Foxp3: Forkhead box protein P3
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
GATA3: GATA-binding protein 3
IFN: Interferon
IL: Interleukin
PVDF: Polyvinylidene fluoride
qRT-PCR: Quantitative real-time polymerase chain reaction
ROR-γt: retinoic acid receptor-related orphan receptor γt
T-bet: T-box expressed in T cell
TNF: Transforming growth factor
TUNEL: Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
SOD: Super Oxide Dismutase
MDA: Malondialdehyde
ACKNOWLEDGMENT
Not applicable.
AUTHOR CONTRIBUTIONS
HZ and YX: Designed the study; all authors conducted the study; HZ and XYC: Collected and analyzed the data; HZ and XYC: Participated in drafting the manuscript, and all authors contributed to critical revision of the manuscript for important intellectual content. All authors gave final approval of the version to be published. All authors participated fully in the work, took public responsibility for appropriate portions of the content, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or completeness of any part of the work were appropriately investigated and resolved.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Experimental Animal Welfare Ethics Committee of Beijing Medical Association (Institutional Review Committee No. MDKN-2024-009). This article does not include patients, so informed consent is not required.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING Not applicable.
References
- Hepatocellular carcinoma: Molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. 2023;42:629-52.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatocellular carcinoma: A narrative review on current knowledge and future prospects. Curr Treat Options Oncol. 2023;24:711-24.
- [CrossRef] [PubMed] [Google Scholar]
- In silico transcriptomic analysis of the chloride intracellular channels (CLIC) interactome identifies a molecular panel of seven prognostic markers in patients with pancreatic ductal adenocarcinoma. Curr Genomics. 2020;21:119-27.
- [CrossRef] [PubMed] [Google Scholar]
- Biophysical characterization of chloride intracellular channel 6 (CLIC6) J Biol Chem. 2023;299:105349.
- [CrossRef] [PubMed] [Google Scholar]
- Applying machine learning algorithms to develop a survival prediction model for lung adenocarcinoma based on genes related to fatty acid metabolism. Front Pharmacol. 2023;14:1260742.
- [CrossRef] [PubMed] [Google Scholar]
- Plasma cell signatures predict prognosis and treatment efficacy for lung adenocarcinoma. Cell Oncol (Dordr). 2024;47:555-71.
- [CrossRef] [PubMed] [Google Scholar]
- Triterpene acids from Rosa roxburghii Tratt fruits exert anti-hepatocellular carcinoma activity via ROS/JNK signaling pathway-mediated cell cycle arrest and mitochondrial apoptosis. Phytomedicine. 2023;119:154960.
- [CrossRef] [PubMed] [Google Scholar]
- Pseudolaric acid B triggers cell apoptosis by activating AMPK/JNK/DRP1/mitochondrial fission pathway in hepatocellular carcinoma. Toxicology. 2023;493:153556.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of chloride intracellular channels as prognostic factors correlated with immune infiltration in hepatocellular carcinoma using bioinformatics analysis. Medicine (Baltimore). 2021;100:e27739.
- [CrossRef] [PubMed] [Google Scholar]
- Cucurbitacin B inhibits non-small cell lung cancer in vivo and in vitro by triggering TLR4/NLRP3/GSDMD-dependent pyroptosis. Pharmacol Res. 2021;170:105748.
- [CrossRef] [PubMed] [Google Scholar]
- Construction of miRNA-mRNA-TF regulatory network for diagnosis of gastric cancer. Biomed Res Int. 2021;2021:9121478.
- [CrossRef] [PubMed] [Google Scholar]
- Helicobacter pylori and antrum erosion-specific gene expression patterns: The discriminative role of CXCL13 and VCAM1 transcripts. Helicobacter. 2008;13:112-26.
- [CrossRef] [PubMed] [Google Scholar]
- IL-17A functions and the therapeutic use of IL-17A and IL-17RA targeted antibodies for cancer treatment. Int Immunopharmacol. 2023;123:110757.
- [CrossRef] [PubMed] [Google Scholar]
- Pro-and anti-inflammatory bioactive lipids imbalance contributes to the pathobiology of autoimmune diseases. Eur J Clin Nutr. 2023;77:637-51.
- [CrossRef] [PubMed] [Google Scholar]
- PKM2 promotes cell metastasis and inhibits autophagy via the JAK/STAT3 pathway in hepatocellular carcinoma. Mol Cell Biochem. 2021;476:2001-10.
- [CrossRef] [PubMed] [Google Scholar]
- Proteomic landscape of the human choroid-retinal pigment epithelial complex. JAMA Ophthalmol. 2014;132:1271-81.
- [CrossRef] [PubMed] [Google Scholar]
- CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898-909.
- [CrossRef] [PubMed] [Google Scholar]








