Translate this page into:
Deciphering the synergistic role of tyrosyl-tRNA synthetase 1 and yes-associated protein 1: Catalysts of malignant progression in hepatocellular carcinoma

*Corresponding author: Lifang Zhou, Department of Hepatobiliary Surgery, Cangzhou Hospital of Integrated TCM- WM•Hebei, Cangzhou, China. zhoulifang2023@163.com
-
Received: ,
Accepted: ,
How to cite this article: Zhou L, Zhang X, Zhang C, Wang Y, Zhang J, Wang Y, Sui Y, et al. Deciphering the synergistic role of tyrosyl-tRNA synthetase 1 and yes-associated protein 1: Catalysts of malignant progression in hepatocellular carcinoma. CytoJournal. 2024;21:66. doi: 10.25259/Cytojournal_91_2024
Abstract
Objective:
Hepatocellular carcinoma (HCC) represents a severe and aggressive malignancy with a poor prognosis, characterized by high incidences of illness and death, making it a critical issue for global health. Tyrosyl-tRNA synthetase 1 (YARS1) is known to be upregulated across various cancers and is considerably linked to tumorigenesis. However, the detailed functions and molecular mechanisms of YARS1 in HCC remain unclear. This research explores the expression of YARS1 in HCC and its role in promoting tumor progression through the yes-associated protein 1 (YAP1) pathway.
Material and Methods:
The potential role and diagnostic significance of YARS1 and YAP1 in HCC were analyzed using relevant datasets. Subsequently, we constructed HCC cell lines with stable knockdown or overexpression of YARS1. In vitro, we used Cell Counting Kit-8 and colony formation assays to examine cell proliferation, terminal deoxynucleotidyl transferase dUTP nick end labeling assays to detect apoptosis, and Transwell migration and invasion assays to assess cell metastasis. Western blotting was employed to analyze the molecular mechanisms. Finally, we developed a lung metastasis model for HCC to assess the impact of YARS1 and YAP1 on tumor spread in a living organism, as well as their interrelationship.
Results:
The findings revealed a notable increase in YARS1 expression in HCC tumors, associated with a worse prognosis. In vitro, YARS1 overexpression significantly increased HCC cell proliferation and metastasis when reducing apoptosis (P < 0.001). In addition, YARS1 overexpression accelerated HCC growth in vivo. Further experiments demonstrated that silencing YAP1 effectively reversed the effects of YARS1 on HCC cell invasion (P < 0.01), apoptosis inhibition (P < 0.01), and metastasis (P < 0.001).
Conclusion:
In summary, this research reveals that YARS1 enhances the malignant progression of HCC through the activation of the YAP1 signaling pathway. Elevated levels of YARS1 in HCC are strongly linked to poor prognosis, indicating that YARS1 might serve as a new therapeutic target for HCC. Future studies should investigate additional mechanisms of YARS1 in HCC and create targeted therapies to improve outcomes for HCC patients.
Keywords
Tyrosyl-tRNA synthetase 1
Hepatocellular carcinoma
Yes-associated protein 1 signaling pathway
Tumor progression
Metastasis
INTRODUCTION
Hepatocellular carcinoma (HCC), characterized by high rates of occurrence and death, is a prevalent cancer globally.[1,2] According to global cancer statistics, HCC ranks as the fourth leading cause of cancer-related mortality worldwide, with its incidence rising in multiple countries.[3,4] The high incidence and mortality of HCC are closely related to its complex pathological mechanisms, difficulties in early diagnosis, and resistance to existing treatments.[5] Hence, research on the molecular pathways of HCC is imperative to discover novel targets for treatment and create successful therapeutic approaches.[6]
Tyrosyl-tRNA synthetase 1 (YARS1) is a ribosomal small subunit homolog whose oncogenic role in various cancers has been gradually revealed in recent years.[7-9]Research has indicated that YARS1 is abundantly present in different types of cancer cells and is strongly linked to the development and advancement of tumors. For example, Wang et al. reported high expression of YARS1 in cancer cells,[10] whereas Zhang et al. found that YARS1 is overexpressed in gastric cancer and can potentially act as a prognostic biomarker.[11] The Hippo pathway plays a vital role in controlling the size of organs and maintaining tissue balance, with its disruption being strongly linked to the formation and progression of different types of cancer.[12] It also manages its biological roles by managing the function of its downstream effector yes-associated protein 1 (YAP1). YAP1 has the ability to move to the nucleus, attach to the transcription factor transcriptional enhanced associate domain, and enhance the transcription of genes that are located downstream, ultimately aiding in the proliferation, invasion, and spread of tumor cells.[13-15]Thus, the YAP1 expression level can be indicative of the Hippo pathway’s activity and the severity of tumors.
While the oncogenic function of YARS1 has been identified in different types of cancers, its precise role and molecular pathways in HCC are still vague. Specifically, the lack of systematic research on whether YARS1 is involved in HCC through the regulation of the Hippo/YAP1 pathway is an important concern. Furthermore, the correlation between YARS1 and the outcome of patients with HCC has yet to be completely confirmed.[16] Thus, investigating the presence of YARS1 in HCC and its impact on prognosis, as well as delving into the molecular pathways through which YARS1 enhances HCC advancement via the YAP1 pathway, are crucial for comprehending the development of HCC and discovering potential treatment targets.[17]
This research focused on examining the presence of YARS1 in HCC and its impact on prognosis, as well as uncovering the molecular processes through which YARS1 enhances HCC advancement by activating the YAP1 pathway. We investigated YARS1 expression levels in HCC tumors and explored their association with unfavorable prognosis by analyzing relevant datasets. Subsequently, we generated HCC cell cultures with consistent reduction or increase of YARS1 and thoroughly examined the impact of YARS1 on HCC cell growth, cell death, movement, and penetration by conducting various experiments inside and outside the body. The study methodically uncovers the cancer-causing function and molecular processes of YARS1 in HCC, revealing that YARS1 accelerates HCC advancement through the activation of the YAP1 pathway. The discovery not only offers fresh perspectives on the molecular processes of HCC but also presents possible new avenues for targeted therapy against HCC. Additional studies are required to investigate the precise regulatory network of YARS1 in HCC and create focused therapeutic approaches targeting YARS1 and its associated pathways to enhance treatment options for patients with HCC.
MATERIAL AND METHODS
Dataset analysis
YARS1 expression in HCC was analyzed using the TNMplot (https://tnmplot.com/analysis/) and Sangerbox 3.0 datasets. The prognosis of YARS1 in HCC was detected using Kaplan– Meier (KM) plotter (https://kmplot.com/analysis/).
Immunohistochemistry (IHC) assays
Fresh HCC tumors ware obtained from patients with HCC and subsequently preserved in liquid nitrogen. This study informed consent from patients was secured, and was strictly adheto the Helsinki Declaration was strictly adhered. The median age of patients was 52 years, with 18 men and six women. The following were the requirements for inclusion: (i) patients having a postoperative clinical diagnosis of liver cancer; (ii) all patients had R0 resection according to histologic investigations; and (iii) matched normal tissue was located <2 cm from tumor tissue. The following were the exclusion criteria: (i) individuals with distant metastases and (ii) those who underwent chemotherapy or radiation therapy before surgery. The clinical sample study was approved by the Committee of Cangzhou Hospital of Integrated TCM- WM Hebei Hospital, approval no. 2023-ky-112.3. For IHC, the samples were initially subjected to incubation with primary antibodies targeting YARS1 (1:200, Z30014, Beijing Baiao Laibo Technology Co., Ltd., Beijing, China) overnight at a temperature of 4°C. Subsequently, we incubated the samples with the second antibody (1:500, RGAM001, Proteintech, Wuhan, China) at room temperature for 1 h. The positive results were detected using DAB and analyzed using an Olympus microscope BX46 (Japan). The parts were given the following proportion scores: 1, 0–1%; 2, 2–10%; 3, 11–30%; 4, 31–70%; and 5, 71–100%. On a scale from 0 to 3, the staining intensity was assessed as follows: 0, negative; 1, weak; 2, moderate; and 3, high. The total score (range, 0–8) was then calculated by summing the proportion and intensity ratings. There are three experts make the IHCs core.
Cell line culture
We obtained Hep3B (HB-8064) and Huh-7 (HB-8065) cell lines from ATCC and maintained them in Dulbecco’s Modified Eagle Medium (DMEM) (C0891, Beyotime, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (C0251, Beyotime, Shanghai, China) and 1% penicillin–streptomycin (C0222, Beyotime, Shanghai, China). The cells were cultured at 37°C in a 5% carbon dioxide (CO2) atmosphere. Every 2–3 days, cell growth was monitored, and when cells reached 80–90% confluence, they were gently washed twice with phosphate buffered saline (PBS).Cells were then incubated with 0.25% trypsin solution (SD1083, Beyotime, Shanghai, China) until they rounded up and detached. Fresh culture medium containing 10% FBS and trypsin was added to detach the cells completely. Cells were resuspended in fresh medium and subcultured into new culture flasks or plates as needed. All cell lines used were authenticated through short tandem repeat profiling and tested negative for mycoplasma contamination.
Cell transfection
pcDNA3.1 (negative control to pcDNA 3.1-YARS1) and pcDNA3.1-YARS1 were generated by GeneChem (Shanghai, China). siNC (negative control to YARS1 siRNA), siYARS1 (YARS1 siRNA), shRNA NC (negative control to YAP1 shRNA), and shRNA YAP1 (5′-CCCAGTTAAATGTTCACCAAT-3′) were purchased from RiboBio. The interference sequence for YARS1 was: 5′-ACTGAACAAGTTGCTGGAT-3′. The standard protocol was followed to transfect the indicated cells with Lipofectamine 2000 (Invitrogen, Waltham, Massachusetts, USA). Puromycin was used to screen YARS1 stably expressed HCC cells.
Cell Counting Kit-8 (CCK-8) assays
Hep3B and Huh-7 cells were grown to logarithmic growth phase and treated according to the experimental design. CCK-8 reagent solution (C0037, Beyotime, Shanghai, China) was prepared and added to the treated cell culture medium. The culture dishes were placed back to the incubator and stored for 2 h at 37°C with 5% CO2. After incubation, the absorbance was measured at 450 nm wavelength using a microplate reader (Synergy H1, Biotek, Winooski, Vermont, USA).
Colony formation assays
Initially, the cells were dispersed into single-cell suspension, ensuring an appropriate number of cells per well (1 × 103 cells/well). These cells were evenly distributed into culture dishes containing suitable growth medium and then incubated at 37°C with 5% CO2 for 7 days, allowing the cells to grow freely and forming colonies. After incubation, the cells were fixed and stained (crystal violet, G1062, Beyotime Biotechnology, Shanghai, China) for 10 min, typically using polyformaldehyde (P1110, Beyotime Biotechnology, Shanghai, China) for fixation, followed by colony counting using a microscope (BX53, Olympus, Tokyo, Japan) on the stained culture dishes.
Cell migration and invasion assays
According to the standard protocol, Transwell permeability support (Corning, New York, USA) with or without Matrix was used to test cell migration and invasion. The upper culture chamber contained cell cultured in serum-free DMEM medium, whereas the bottom chamber was filled with DMEM medium supplemented with 10% FBS. After 48 h of incubation, the cells in the upper chamber were scraped off, stained with crystal violet for 20 min (G1062, Beyotime Biotechnology, Shanghai, China), and counted.
Western blot assay
Radioimmunoprecipitation assay lysis buffer (P0013B, Beyotime, Shanghai, China) was used to extract proteins from cells, and the Bicinchoninic Acid Assay method was applied to determine the protein concentration. Using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel electrophoresis, the extracted protein samples were separated. Using a polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany), proteins that had been isolated in the SDS-PAGE gel were introduced. To stop nonspecific binding, 5% bovine serum albumin was used to block the membrane after transfer. Specific primary antibodies (Anti-YARS1 [1:500, PA5-53883, Abcam, Cambridge, MA, USA]), YAP1 [1:1000, ab205270, Abcam, Cambridge, MA, USA], Bax [1:1000, #5023, CST, Ma, BSN, USA], and Bcl-2 [1:1000, #4223, CST, Ma, BSN, USA], were used to recognize the target protein and incubated overnight at room temperature. Unbound primary antibodies were removed using wash buffer containing Tween-20. A secondary antibody (1:1000, ab6728, Abcam, Cambridge, MA, USA) binding to the primary antibody was incubated with the membrane and then washed to remove unbound secondary antibody. Chemiluminescent reagents (A38554, Thermo Fisher Scientific, Waltham, Massachusetts, USA) were used to detect signals of specific proteins on the membrane, employing the gel imaging system (ChemiDoc, Bio-Rad Laboratories, Hulsey, California, USA). Quantitative analysis of protein expression levels was performed using ImageJ software (version 1.5e, National Institutes of Healthm, Bethesda, Maryland, USA), considering the developed membrane image after exposure.
Animals
Forty-eight male specific pathogen free grade nude mice, aged 6 weeks and weighing between 15 and 20 g, were acquired from Shanghai Slack Laboratory Animal Co., Ltd. (animal production license number: SYXK [Shanghai] 2020– 0009). Animal experimentation in this study was approved by the Committee of Cangzhou Hospital of Integrated TCM- WM•Hebei under approval no. czx2023-ky-101. The experiment commenced after a week of acclimatization. All mice maintained on a 12 h light–dark cycle at 23 ± 1°C and supplied free food and water. Cells in the logarithmic growth phase were detached using 0.25% trypsin to create a cell suspension with a concentration of 2 × 107 cells/mL. The mice were divided into control (Hep3B cells or Huh-7 cells transfected with pcDNA3.1), YARS1 (Hep3B cells or Huh-7 cells transfected with pcDNA3.1-YARS1), NC (Hep3B cells transfected with shRNA NC), and shYARS1 (Hep3B cells transfected with shRNA YAP1). This suspension was injected into the larger tail veins of the nude mice,[18] with each mouse receiving approximately 0.5 mL. Six weeks after the injection, the mice were euthanized by administering an intraperitoneal injection of sodium pentobarbital (57-33-0, Sigma-Aldrich, St. Louis, Missouri, USA) at a dose of 110 mg/kg. After injection through the tail vein, tumor cells initially enter the arterial blood circulation system through the pulmonary capillary network, causing multiple metastases throughout the body. However, because tumor cells are sticky and easy to cluster, they are generally trapped in the micro-vessels of the mouse lung, mainly forming lung metastasis.[19] Lung tissues were harvested, assessed for metastases, and then fixed in 10% formaldehyde for 24 h. The tissues were dehydrated using a series of ethanol concentrations (80%, 90%, and 100%) and butanol, embedded in paraffin at 60°C, sectioned, and stained with hematoxylin and eosin (G1120, Solarbio, Beijing, China) for examination.
Hematoxylin–eosin (HE) staining
Pathological changes in the metastatic foci were examined using hematoxylin–eosin staining. Lung metastatic foci from the mice were fixed in 4% formaldehyde for 15 min and then dehydrated through a series of graded ethanol (100%, 95%, 85%, and 75%) for 3 min each. The tissue was embedded in paraffin and sectioned using a microtome (RM2235, Leica, Wetzlar, Baden-Württemberg, Germany) (5–8 mm). The tissue sections were immersed in hematoxylin stain for 5 min, differentiated in acid and ammonia water for 30 s each, and blued under running water for 5 min, dehydrated in 70% and 90% ethanol for 10 min, incubated in eosin for 5 min, and dehydrated in absolute ethanol for 2 min. After clearing in xylene for 5 min, the sections were quickly air-dried and mounted for microscopic examination using a BX53 microscope (Olympus, Tokyo, Japan).
Immunofluorescence for E-cadherin and vimentin expression
Triton X-100 was used to permeabilize the treated cells, and goat serum was then used to block them for an hour at room temperature. The cells were treated with the appropriate primary antibodies for a whole night at 4°C after being washed with PBS. Primary antibodies include E-Cadherin (4A2) Mouse #14472 (1:200) and Vimentin (D21H3) Rabbit mAb #5741 (1:200). They were purchased from CST, Ma, BSN, USA. Following PBS washes, the cells were incubated with fluorescent secondary antibodies (ab150127 or ab7050, 1:1000, Abcam, Cambridge, MA, USA) in the dark at room temperature for 1 h. The cells were then stained with 4›,6-diamidino-2-phenylindole (DAPI) (1 mg/mL) for 15 min in the dark. After a final PBS wash, an anti-fluorescence quenching mounting medium was applied. Cell morphology was captured using a laser scanning confocal microscope (Leica SP-8, Wetzlar, Switzerland). Protein expression was evaluated by analyzing the average fluorescence intensity using ImageJ software (version 1.5e, National Institutes of Health, Bethesda, Maryland, USA).
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay for apoptosis detection
Glass coverslips were soaked overnight in anhydrous ethanol and placed in six-well plates, and 1 × 105 cells were seeded per well. When the cells reached the logarithmic growth phase, they were transfected. Following 48 h of transfection, the TUNEL assay was carried out in accordance with the guidelines provided by the TUNEL BrightGreen Apoptosis Detection Kit (A112-01, Vazyme, Nanjing, China). For 25 min, the cells were fixed in 4% paraformaldehyde. After soaking Triton X-100 for 10 min, the Triton X-100 was stained with 50 mL TdT solution and 450 mL fluorescein-labeled dUTP solution for 60 min in the dark. Then, DAPI was stained for 15 min. Under a confocal laser scanning microscope (LSM 880, Zeiss, Oberkochen, Baden–Württemberg, Germany), green fluorescence was observed at 488 nm, and blue fluorescence at 405 nm. Cells showing green fluorescence in the nucleus were considered apoptotic.
Statistical methods
Experimental data were statistically analyzed and visualized using GraphPad Prism 8.0 software (GraphPad Software, Inc., San Diego, California, USA). Quantitative data were presented as mean ± standard deviation. Normality and homogeneity of variance were assessed using these data. For normally distributed data, one-way analysis of variance was performed for multiple group comparisons, followed by the Least Significant Difference test for pairwise comparisons. For data that did not satisfy the assumptions of normality or homogeneity of variance, the Kruskal–Wallis test was used for multiple group comparisons, and the t-test was performed for pairwise comparisons. A P-value below 0.05 was deemed to indicate statistical significance.
RESULTS
YARS1 is overexpressed in HCC
We analyzed the expression profile of YARS1 in the The Cancer Genome Atlas dataset online to understand its role in HCC. The result showed that YARS1 was overexpressed in the HCC tumor [Figure 1a]. Furthermore, we confirmed that YARS1 was positively correlated with poor prognosis using the KM mapping database [Figure 1b]. Analysis results from the Sangerbox 3.0 database also indicated that YARS1 was positively correlated with poor prognosis [Figure 1c]. To further verify the expression of YARS1 in HCC, we conducted IHC and HE assays and confirmed that YARS1 was elevated in tumor (P < 0.001) [Figure 1d-f]. These data suggested that YARS1 expression was associated with HCC progression.
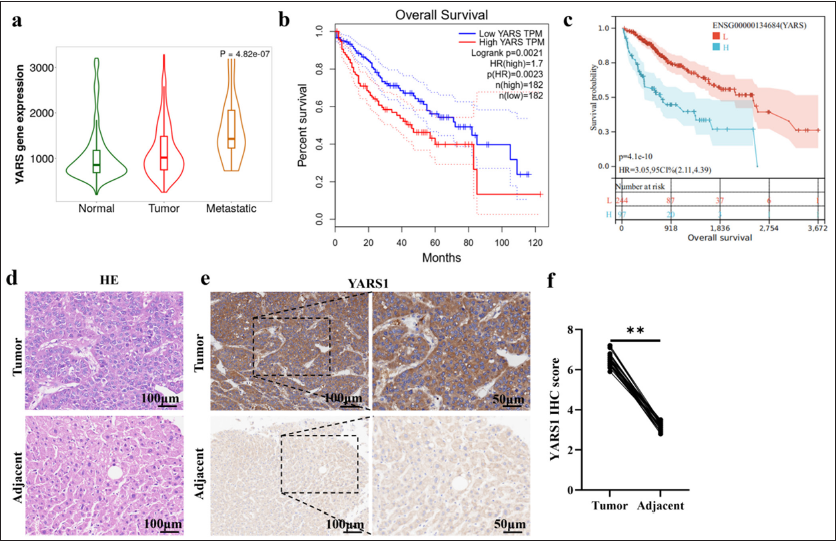
- Tyrosyl-tRNA synthetase 1 (YARS1) is overexpressed in hepatocellular carcinoma (HCC). (a) YARS1 expression in HCC tumors. (b and c) The relevance of YARS1 levels and overall survival curves in HCC. (d) Representative photographs of YARS1 hematoxylin–eosin assays in HCC tumors. (e) Representative photographs of YARS1 immunohistochemistry (IHC) assays in HCC tumors. (f) Statistical quantification of IHC score. ✶✶✶ P < 0.001.
YARS1 enhances cell growth of HCC in vitro
YARS1 function in promoting HCC cell growth was examined by transfecting YARS1 siRNA and YARS1 overexpression plasmids into Hep3B and Huh-7 cells. Then, transfection efficiency was assessed using Western blot assays. The findings indicated a significant decrease in YARS1 expression in Hep3B and Huh-7 cells following transfection with YARS1 siRNA (P < 0.001). By contrast, YARS1 expression increased in Hep3B and Huh-7 cells after transfection with a YARS1 overexpression plasmid (P < 0.01) [Figure 2a-f]. Furthermore, we evaluated the role of YARS1 on cell growth and found that reducing YARS1 in Hep3B and Huh-7 cells notably decreased cell proliferation. On the contrary, increasing YARS1 resulted in a significant enhancement in cell proliferation (P < 0.001) [Figure 2g-r]. These findings demonstrated that YARS1 could promote HCC cell growth in vitro.
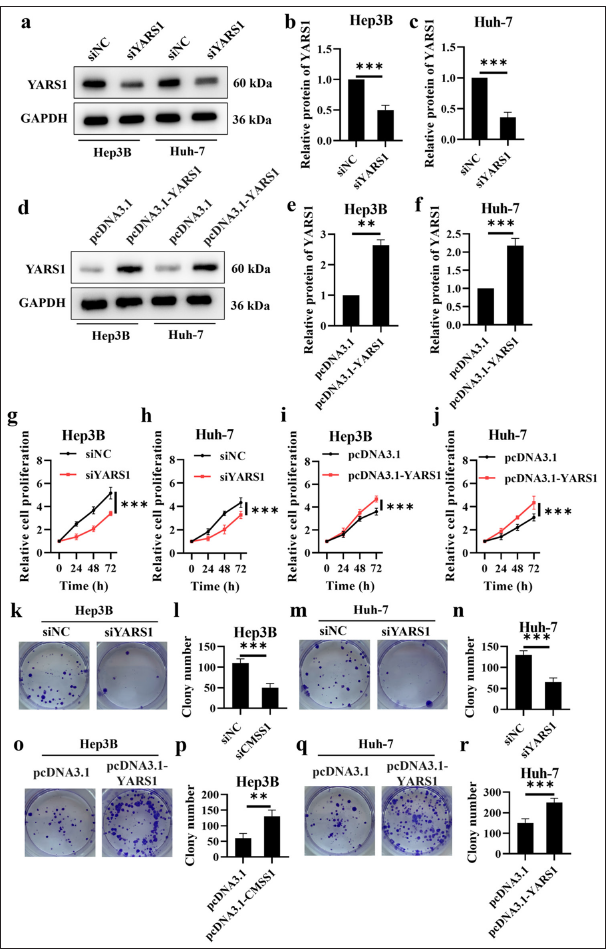
- Tyrosyl-tRNA synthetase 1 (YARS1) enhances cell proliferation of Hepatocellular carcinoma in vitro. (a-f) Western blot analysis was performed on YARS1 in Hep3B and Huh-7 cells following transfection with specified plasmids. (g-j) Proliferation rate of Hep3B and Huh-7 cell transfection of indicated plasmids. (k-r) Representative photographs and statistical quantification of colony in Hep3B and Huh-7 cell transfection of indicated plasmids. n = 3. ✶✶ P < 0.01, ✶✶✶ P < 0.001. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
YARS1 inhibits cell apoptosis of HCC in vitro
The impact of YARS1 on programmed cell death in Hep3B and Huh-7 cells was investigated through TUNEL assays. The result showed that YARS1 overexpression decreased the apoptosis rate (P < 0.001) [Figure 3a-d]. Moreover, YARS1 overexpression inhibited bax expression, but induced a significant upregulation of Bcl-2 expression (P < 0.001) [Figure 3e-i]. This finding suggested that YARS1 can effectively inhibit apoptosis in HCC cells.
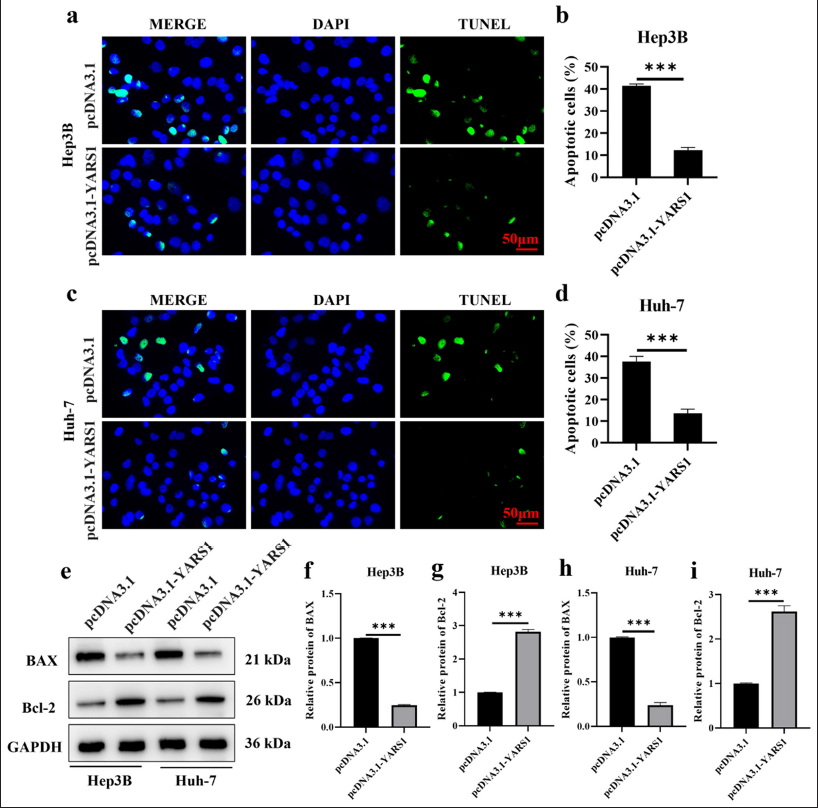
- Tyrosyl-tRNA synthetase 1 inhibits cell apoptosis of Hepatocellular carcinoma in vitro. (a-d) Representative photographs and statistical quantification of apoptosis of Hep3B and Huh-7 cell transfection of indicated plasmids. (e-i) Western blot tests were conducted on bax and Bcl-2 proteins in Hep3B and Huh-7 cells after transfection with specified plasmids. n = 3. ✶✶✶ P < 0.001. DAPI: 4’,6-diamidino-2-phenylindole, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, Bcl-2: B-cell lymphoma 2, Bax: BCL2-associated X protein, TUNEL: Terminal deoxynucleotidyl transferase dUTP nick end labeling.
YARS1 promoted HCC migration and invasion
The migration test measured cell movement, whereas the invasion test involved cells releasing proteases to break down the extracellular matrix, expanding the results of the migration test.[20] Both tests were crucial for evaluating the cancerous nature of tumor cells. The findings indicated a notable increase in the number of Hep3B and Huh-7 cells with elevated YARS1 levels crossing the Transwell chamber membrane compared with the control group. This study demonstrated that increased YARS1 expression significantly enhanced the migratory (P < 0.01) and invasive (P < 0.001) capabilities of HCC cells [Figure 4a-f]. This observation suggested that YARS1 may play a role in regulating the motility and protease secretion of HCC cells. Immunofluorescence double staining was used to assess changes in E-cadherin and vimentin expression, markers associated with epithelial–mesenchymal transition (EMT). Our findings indicated that overexpression of YARS1 reduced E-cadherin and increased vimentin expression in Hep3B and Huh-7 cells [Figure 4g and h]. These findings suggested that YARS1 promoted the EMT process in HCC cells, thereby enhancing their migration and invasion capabilities. Furthermore, we assessed the impact of YARS1 on in vivo metastasis by injecting tumor cells into the tail veins of mice. The number of lung metastases reflected the in vivo metastatic potential of tumor cells, given that the lungs were a common site of hematogenous metastasis in HCC. HE staining revealed a greater number of lung metastatic foci in mice overexpressing YARS1 than the control group (P < 0.001) [Figure 4i-l], indicating that YARS1 overexpression enhances the in vivo metastasis of HCC cells.
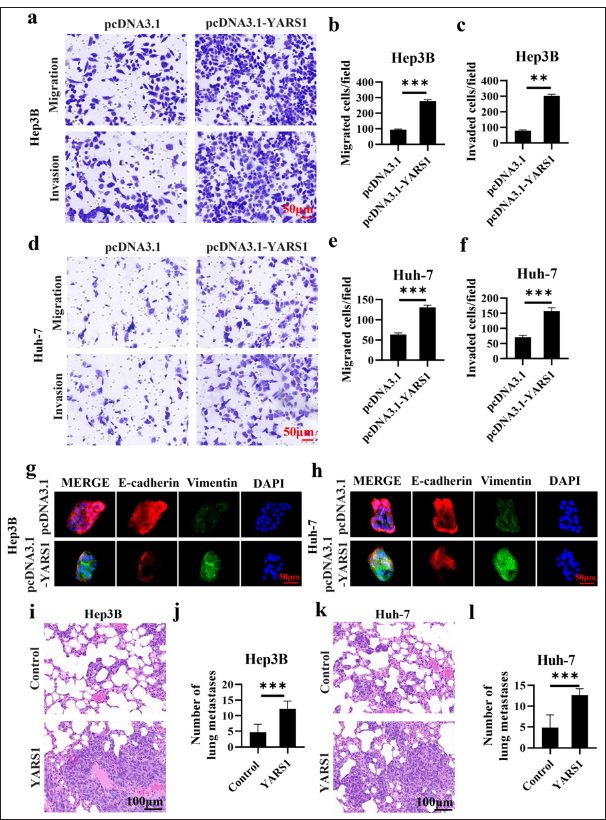
- Tyrosyl-tRNA synthetase 1 (YARS1) promoted migration and invasion of Hepatocellular carcinoma. (a-f) Representative photographs and statistical quantification of migration and invasion of indicated plasmids. (g and h) E-cadherin and vimentin in Hep3B and Huh-7 cell transfection of indicated plasmids. (i-l) After YARS1 overexpression, Hep3B and Huh-7 tumor cells were injected via the tail vein, and the ability to form lung metastases was assessed by hematoxylin–eosin staining. n = 3. ✶✶ P < 0.01, ✶✶✶ P < 0.001.
YARS1 increased YAP1 activity
YAP1 plays a critical role as a mediator within the Hippo pathway and is crucial for tumor development and progression.[21] Therefore, the level of YAP1 protein serves as an indicator of Hippo pathway activity and tumor severity. In this study, we elevated YARS1 expression in two HCC cell lines, Hep3B and Huh-7, using an empty vector as a control (pcDNA3.1). We also assessed changes in YAP1 protein levels via Western blot analysis. Our findings revealed a significant increase in YAP1 protein expression in Hep3B (P < 0.05) and Huh-7 (P < 0.01) cells following YARS1 overexpression [Figure 5a-c].
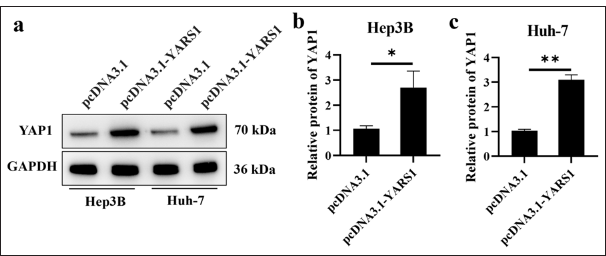
- Tyrosyl-tRNA synthetase 1 increased yes-associated protein 1 (YAP1) activity. (a-c) Western blot tests were conducted on YAP1 in Hep3B and Huh-7 cells after transfection with specified plasmids. n = 3. ✶ P < 0.05, ✶✶ P < 0.01. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
YARS1 promoted HCC growth
The main focus of this research was to determine if inhibiting YAP1 could counteract the cancer-causing impacts of YARS1 to confirm the role of YARS1 in promoting the advancement of HCC via the YAP1 pathway. Western blot analysis indicated a notable increase in YARS1 protein expression following YARS1 overexpression. YAP1 suppression in the presence of YARS1 upregulation led to a substantial decrease in YAP1 protein levels (P < 0.001), whereas the control group treated with scrambled shRNA did not exhibit any notable impact. This finding indicates that the shRNA efficiently interfered with YAP1 expression [Figure 6a-c]. The Transwell invasion assay showed a notable increase in invading cells in the YARS1 overexpression group, indicating that YARS1 enhances the invasion of HCC cells. Silencing YAP1 and overexpressing YARS1 significant decreased the number of invading cells (P < 0.01), similar to the control group. This result suggested that inhibiting YAP1 can successfully counteract the enhancing impact of YARS1 on invasiveness [Figure 6d and e]. The TUNEL assay findings indicated a notable decrease in the quantity of TUNEL-positive cells in the YARS1 overexpression group (P < 0.01), indicating that YARS1 suppressed apoptosis in HCC cells. Silencing YAP1 when overexpressing YARS1 notably increased the amount of TUNEL-positive cells (P < 0.01), similar to the control group. This outcome indicated that silencing YAP1 can effectively reverse the inhibitory effect of YARS1 on cell apoptosis [Figure 6f and g]. The tumor cell metastatic potential was evaluated by establishing a lung metastasis model in nude mice by injecting tumor cells into the tail vein. The findings indicated a notable increase in lung metastases in the YARS1 overexpression group (P < 0.001), indicating the role of YARS1 in promoting HCC cell metastasis. Silencing YAP1 when overexpressing YARS1 considerably reduced the quantity of lung metastases. This finding suggested that inhibiting YAP1 can successfully counteract the enhancing impact of YARS1 on metastasis [Figure 6h and i]. This study used various functional experiments, such as Western blot, Transwell invasion, TUNEL apoptosis, and nude mouse lung metastasis, to systematically demonstrate that inhibiting YAP1 can successfully counteract the enhancing impacts of YARS1 on HCC cell invasion, apoptosis suppression, and metastasis. This result further confirmed that YARS1 enhanced the advancement of HCC by activating the YAP1 pathway.
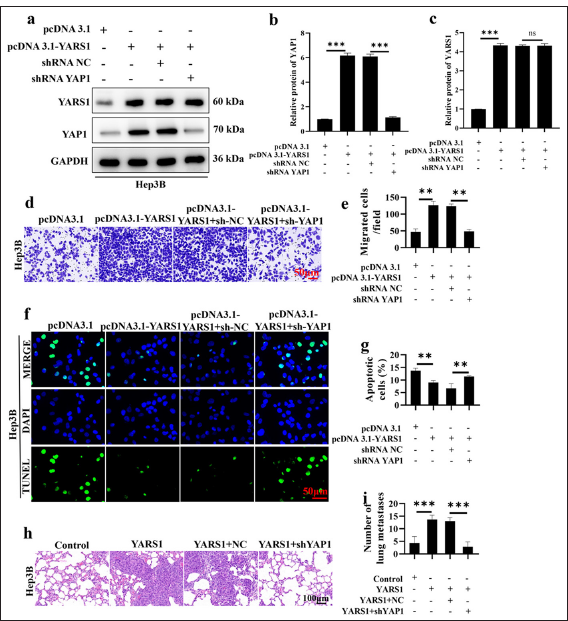
- Silencing yes-associated protein 1 (YAP1) reverses the tumor-promoting effect of Tyrosyl-tRNA synthetase 1 (YARS1). (a-c) Western blot was conducted to verify the changes in YARS1 and YAP1 expression levels in different treatment groups. (d and e) Transwell assay was performed to examine the effect of YAP1 silencing on the tumor-promoting function of YARS1. (f and g) Terminal deoxynucleotidyl transferase dUTP nick end labeling assay to verify the effect of YAP1 silencing on the ability of YARS1 to regulate cell apoptosis. (h and i) Six weeks after injecting cells into the tail vein of nude mice, lung metastasis was detected. n = 3. ✶✶ P < 0.01, ✶✶✶ P < 0.001. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, pcDNA 3.1: Negative control to pcDNA 3.1-YARS1, shRNA NC: Negative control to YAP1 shRNA, NC: Hep3B cells transfected with shRNA NC.
DISCUSSION
HCC is a prevalent cancer globally, posing a significant health challenge due to its high rates of occurrence and death.[3,22] Despite advancements in diagnosing and treating HCC, the outlook for patients is still grim because of its aggressive and spreading characteristics.[23] Thus, delving into the molecular pathways of HCC and discovering novel treatment targets hold substantial importance in clinical and scientific realms.
High levels of YARS1 have been detected in different types of cancers, indicating a strong link to the development and advancement of tumors.[10,11,16,24] This research methodically uncovered the cancer-causing function of YARS1 in HCC and its molecular process in enhancing HCC aggressiveness by activating the YAP1 pathway in various laboratory and animal tests. The dataset analysis revealed that YARS1 is upregulated in HCC tumor tissues and that its elevated expression is associated with a poor prognosis in patients. This discovery indicates that YARS1 could remarkably affect the onset and progression of HCC. To confirm this theory, we developed HCC cell cultures with consistent reduction and increase in YARS1 and carried out several practical tests. YARS1 overexpression in in vitro studies was found to greatly enhance the growth, movement, and infiltration of HCC cells, while also suppressing cell death. YARS1 overexpression notably enhanced the proliferative ability of HCC cells, as demonstrated by CCK8 and colony formation assay results. Moreover, the overexpression of YARS1 significantly inhibited apoptosis in HCC cells. Transwell assays showed that increased expression of YARS1 significantly boosted the migration and invasion abilities of HCC cells. In vivo experiments further validated the oncogenic role of YARS1 in HCC using a subcutaneous tumor model. The results showed that YARS1 overexpression significantly promoted tumor growth. Moreover, YARS1 overexpression notably enhanced the lung metastatic potential of HCC cells in a lung metastasis model created through the injection of tumor cells into the tail vein.
We performed a series of molecular biology experiments to explore the molecular mechanisms through which YARS1 enhances HCC malignancy. The results indicated that YARS1 promotes HCC malignancy by activating YAP1. We carried out experiments to silence YAP1, confirming the theory that YARS1 enhances HCC aggressiveness via the YAP1 pathway. The findings showed that suppressing YAP1 effectively counteracted the impact of increased YARS1 on the growth, movement, infiltration, and cell death of HCC cells, providing additional evidence that YARS1 enhances HCC aggressiveness by activating the YAP1 pathway.
Despite systematically revealing the oncogenic role of YARS1 in HCC and its molecular mechanisms, our study still presents some limitations. YARS1 could be involved in the initiation and progression of HCC through alternative pathways, prompting further investigation into additional mechanisms linking YARS1 to HCC. Our research primarily focused on the role and operation of YARS1. Future research should explore the potential of YARS1 as a treatment target for HCC and create specific treatment plans aimed at YARS1.
SUMMARY
The research methodically uncovered the cancer-causing function of YARS1 in HCC and its molecular process in enhancing HCC aggressiveness through the activation of the YAP1 pathway. YARS1 specifically enhances the growth, movement, and infiltration of HCC cells through the increased expression of YAP1. The overexpression of YARS1 is associated with unfavorable outcomes in individuals with HCC, indicating its potential as a prognostic indicator and treatment target for this condition. Future studies should further explore other mechanisms involving YARS1 in HCC, develop targeted therapeutic strategies against YARS1, and evaluate its clinical therapeutic potential. The study highlights the importance of YARS1 in HCC and provides fresh perspectives on the molecular pathways involved in HCC, suggesting a promising new target for HCC treatment. We expect that future studies will advance the application of YARS1 in HCC treatment, improving the prognosis and survival rates of HCC patients.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study were available from the corresponding author on reasonable request.
ABBREVIATIONS
BSA – Bovine serum albumin
CO2 – Carbon dioxide
DAPI – 4’,6-diamidino-2-phenylindole
DMEM – Dulbecco’s Modified Eagle Medium
FBS – Fetal bovine serum
HCC – Hepatocellular carcinoma
HE – Hematoxylin-eosin
IHC – Immunohistochemistry
PBS – Phosphate buffer saline
pcDNA 3.1 – Negative control to pcDNA 3.1-YARS1
RIPA – Radioimmunoprecipitation assay
SDS-PAGE – Sodium dodecyl sulfate polyacrylamide gel electrophoresis
shRNA NC – Negative control to YAP1
shRNA siNC – Negative control to YARS1 siRNA
SPF – Specific pathogen free
TCGA – The Cancer Genome Atlas
TEAD – Transcriptional enhanced associate domain
TUNEL – Terminal deoxynucleotidyl transferase-mediated
dUTP nick end labeling
YAP1 – Yes-associated protein 1
YARS1 – Tyrosyl-tRNA synthetase 1
AUTHOR CONTRIBUTIONS
LFZ and XZ: Designed the study; all authors conducted the study; CYZ and YW: Collected and analyzed the data; JJZ, YXW, and YBS: Participated in drafting the manuscript, and all authors contributed to critical revision of the manuscript for important intellectual content. All authors gave final approval of the version to be published. All authors participated fully in the work, take public responsibility for appropriate portions of the content, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or completeness of any part of the work are appropriately investigated and resolved.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Clinical sample study has been approved by the Committee of Cangzhou Hospital of Integrated TCM-WM·Hebei, approval no.: 2023-ky-112.3. Ethics date: November 23, 2023. This study has obtained informed consent from patients and strictly adhered to the Helsinki Declaration.
Animal experimentation in this study has been approved by the Committee of Cangzhou Hospital of Integrated TCM-WM·Hebei, approval no.: czx2023-ky-101. Ethics date: October 10, 2023.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151-72.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatocellular carcinoma: New developments. Clin Liver Dis. 2023;27:85-102.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73:4-13.
- [CrossRef] [PubMed] [Google Scholar]
- Management of hepatocellular carcinoma: A review. JAMA Surg. 2023;158:410-20.
- [CrossRef] [PubMed] [Google Scholar]
- Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61.
- [CrossRef] [PubMed] [Google Scholar]
- Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:146.
- [CrossRef] [PubMed] [Google Scholar]
- The recurrent missense mutation p.(Arg367Trp) in YARS1 causes a distinct neurodevelopmental phenotype. J Mol Med (Berl). 2021;99:1755-68.
- [CrossRef] [Google Scholar]
- Novel partial loss-of-function variants in the tyrosyl-tRNA synthetase 1 (YARS1) gene involved in multisystem disease. Eur J Med Genet. 2021;64:104294.
- [CrossRef] [PubMed] [Google Scholar]
- A missense, loss-of-function YARS1 variant in a patient with proximal-predominant motor neuropathy. Cold Spring Harb Mol Case Stud. 2022;8:a006246.
- [CrossRef] [PubMed] [Google Scholar]
- Unveiling the role of YARS1 in bladder cancer: A prognostic biomarker and therapeutic target. J Cell Mol Med. 2024;28:1-20.
- [CrossRef] [Google Scholar]
- YARS as an oncogenic protein that promotes gastric cancer progression through activating PI3K-Akt signaling. J Cancer Res Clin Oncol. 2020;146:329-42.
- [CrossRef] [PubMed] [Google Scholar]
- The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin Sci (Lond). 2022;136:197-222.
- [CrossRef] [PubMed] [Google Scholar]
- Hippo pathway: Regulation, deregulation and potential therapeutic targets in cancer. Cancer Lett. 2021;507:112-23.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nature Rev Drug Discov. 2020;19:480-94.
- [CrossRef] [PubMed] [Google Scholar]
- The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246-57.
- [CrossRef] [PubMed] [Google Scholar]
- Functional and pathologic association of aminoacyl-tRNA synthetases with cancer. Exp Mol Med. 2022;54:553-66.
- [CrossRef] [PubMed] [Google Scholar]
- Multi-Omics database analysis of aminoacyl-tRNA synthetases in cancer. Genes (Basel). 2020;11:1384.
- [CrossRef] [PubMed] [Google Scholar]
- Morphine suppresses liver cancer cell tumor properties in vitro and in vivo. Front Oncol. 2021;11:666446.
- [CrossRef] [PubMed] [Google Scholar]
- Study on the hepatocellular carcinoma model with metastasis. Genes Dis. 2020;7:336-50.
- [CrossRef] [PubMed] [Google Scholar]
- The many faces of metalloproteases: Cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 2001;11:S37-43.
- [CrossRef] [Google Scholar]
- Hippo pathway-mediated YAP1/TAZ inhibition is essential for proper pancreatic endocrine specification and differentiation. Elife. 2024;13:e84532.
- [CrossRef] [PubMed] [Google Scholar]
- The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology. 2023;77:1773-96.
- [CrossRef] [PubMed] [Google Scholar]
- EPHB2 Activates β-catenin to enhance cancer stem cell properties and drive sorafenib resistance in hepatocellular carcinoma. Cancer Res. 2021;81:3229-40.
- [CrossRef] [PubMed] [Google Scholar]
- AminoacyltRNA synthetase-based prognosis model and exploration of potential mechanisms in hepatocellular carcinoma. J Clin Transl Hepatol. 2023;11:877-88.
- [Google Scholar]








