Translate this page into:
Interactions of tumor necrosis factor receptor-associated factor 4 and pyruvate kinase muscle isoform 2 promote malignant behavior and aerobic glycolysis in colorectal cancer cells

*Corresponding author: Yuanyuan Ma, Department of Traditional Chinese Medicine and Anorectal, Jinan Fourth People’s Hospital, Jinan, Shandong, China. mayuanyuan0228@126.com
-
Received: ,
Accepted: ,
How to cite this article: Liu T, Zhu S, Sun J, Ma Y. Interactions of tumor necrosis factor receptor-associated factor 4 and pyruvate kinase muscle isoform 2 promote malignant behavior and aerobic glycolysis in colorectal cancer cells. CytoJournal. 2025;22:24. doi: 10.25259/Cytojournal_167_2024
Abstract
Objective
Colorectal cancer (CRC) is a malignant tumor of the digestive system, and the main causes of death are metastasis and recurrence. Tumor necrosis factor receptor-associated factor 4 (TRAF4) is associated with the development of various tumors, but its role in CRC development is limited, especially glycolysis. Therefore, TRAF4’s role in the regulation of cell malignant behavior and glycolysis and its specific mechanism were explored in CRC.
Material and Methods
The TRAF4 or pyruvate kinase muscle isoform 2 (PKM2) gene expression was inhibited or promoted by short hairpin ribonucleic acid (sh- RNA) or overexpression (oe) plasmids in Lovo cells. Transfection efficiency was detected by Western blot and real-time quantitative polymerase chain reaction. Cell growth and colony formation were assessed using 5-ethynyl-2’-deoxyuridine and clone formation assays, respectively, and cell migration and invasion ability were observed by scratch healing and Transwell assay. Glucose uptake and lactate production were measured with a kit and used in evaluating the glycolysis capacities of the cells. The levels of TRAF4, PKM2, and glycolytic-related and wingless-type (Wnt)/beta (β)-catenin pathway-related proteins were detected by Western blot, and co-immunoprecipitation (Co-IP) verified TRAF4 and PKM2 interaction in CRC cells.
Results
TRAF4 expression increased in CRC cell lines (P < 0.05, P < 0.001, P < 0.0001). After sh-TRAF4, oeTRAF4, or oe-PKM2 transfection, TRAF4 or PKM2 expression levels in the Lovo cells decreased or increased (P < 0.05, P < 0.01, P < 0.001, and P < 0.0001). TRAF4 knockdown inhibited cell malignant behavior, glucose uptake, lactate production, and glucose transporter type 1 (GLUT1), hexokinase 2 (HK2), PKM2, and lactate dehydrogenase A (LDHA) protein expression levels in CRC cells (P < 0.01, P < 0.001, P < 0.0001). Co-IP experiment showed that TRAF4 was bound to PKM2. PKM2 protein level decreased after TRAF4 knockdown (P < 0.0001), and PKM2 protein expression increased when TRAF4 was overexpressed (P < 0.001). PKM2 overexpression offset the effect of TRAF4 knockdown on cell malignant behavior and aerobic glycolysis (P < 0.05, P < 0.01, P < 0.001, and P < 0.0001). Moreover, Wnt/β-catenin pathway proteins were inhibited after TRAF4 knockdown and were restored by PKM2 overexpression (P < 0.01 and P < 0.0001). Notably, the effects of TRAF4 or PKM2 overexpression on cell malignant behavior, glucose uptake, lactate production, and GLUT1, PKM2, HK2, and LDHA protein expression levels were partially offset by the Wnt/β-catenin signaling suppressor XAV939 (P < 0.05, P < 0.01, P < 0.001, and P < 0.0001).
Conclusion
TRAF4 and PKM2 are associated with CRC development. TRAF4 binds to PKM2 and promotes CRC malignant behavior and glycolysis through the Wnt/β-catenin signaling pathway.
Keywords
Aerobic glycolysis
Colorectal cancer
Pyruvate kinase muscle isoform 2
Tumor necrosis factor receptor-associated factor 4
INTRODUCTION
Colorectal cancer (CRC) occurs in colon glandular epithelial cells and easily recurs and metastasizes, resulting in high mortality.[1] It is a heterogeneous disease with an extremely complex pathogenesis that remains not fully understood.[2] The onset of CRC is relatively insidious and sudden.[3] CRC treatment is usually based on surgery and supplemented by chemotherapy and radiotherapy, but the postoperative recurrence rate and fatality rate are high.[4] Hence, exploring the mechanism underlying CRC malignant evolution and novel therapeutic targets is particularly important.
Cell proliferation is an important intrinsic property that maintains homeostasis in the internal environment of the body and maintains a relatively constant number of cell types and cells in various organs, but it causes a variety of diseases once disorders occur.[5] Infinite proliferation is the most important component in tumor progression, and inhibiting tumor cell proliferation is the basic strategy of tumor therapy.[6] The abnormally rapid proliferation of tumor cells is accompanied by abnormally vigorous cell metabolism.[7] Glycolysis is a common metabolic pathway during tumor cell development.[8,9] Compared with normal cells, tumor cells tend to favor the productive mode of glycolysis even under oxygen-abundant conditions.[10,11] Glycolysis activity increases in proliferating cells, and glycolysis inhibition impedes cancer development.[12] Therefore, understanding the regulatory mechanism underlying glycolysis in CRC benefits CRC prevention and treatment.
Tumor necrosis factor receptor-associated factor 4 (TRAF4) acts as a connector molecule for signal transduction and has E3 ubiquitin ligase activity.[13] It has vital roles in neurogenesis, embryonic development, immune regulation, inflammatory response, oxidative damage, and metabolism.[13] TRAF4 has abnormally high expression levels in various tumors and is associated with tumor occurrence and development.[14] TRAF4 is an oncogene that was first identified in breast cancer, and inhibiting TRAF4 impedes cell proliferation.[13,15] Silencing TRAF4 inhibits osteosarcoma cell growth in vivo and in vitro.[16] In addition, TRAF4 promotes lung cancer progression.[17] TRAF4 regulates β-catenin, matrix metalloproteinase-9, and matrix metalloproteinase-2 protein expression and affects the invasion and migration of oral cancer cells.[18] TRAF4 promotes high-grade serous ovarian cancer progression by regulating yes-associated protein signaling.[19] In CRC, TRAF4 is highly expressed and participates in the regulation of chemotherapy drug resistance and radioresistance.[20,21] Knocking down TRAF4 inhibits cell proliferation and invasion, ultimately inhibiting CRC development.[22] However, the role of TRAF4 and its mediated biological function is not fully understood, and reports on the correlation between TRAF4 and the incidence of CRC are few.
Pyruvate kinase muscle isoform 2 (PKM2), as an important rate-limiting enzyme in the glycolysis pathway, promotes disease progression in various types of tumors by affecting the tumor microenvironment.[23] When PKM2 expression is up-regulated, PKM2 replaces original tissue-specific pyruvate kinase muscle isoform 1, pyruvate kinase liver, and pyruvate kinase red blood cells during the development of malignant tumors, and thus, some scientists refer to PKM2 as a tumor-specific pyruvate kinase.[24] PKM2 is dysregulated in tumors and promotes tumor cell glycolysis, which is necessary for cell proliferation, metabolism, invasion, and metastasis.[25,26] High PKM2 expression promotes cell proliferation and migration through multiple signaling pathways and is associated with prognosis in CRC.[27] An analysis of the protein interaction database HitPredict showed that PKM2 can interact with TRAF4. Therefore, we hypothesize that TRAF4 may regulate CRC development through its interaction with PKM2. In summary, this study preliminarily explored the role and possible mechanism of TRAF4 and PKM2 in CRC through in vitro cell experiments to provide a theoretical basis for further exploring the pathogenesis and development of CRC and seek novel therapeutic targets for CRC.
MATERIAL AND METHODS
Cell culture and transfection
NCM460 (normal colorectal epithelial cell line; BNCC339288, Bena, Jiangsu, China) and CRC cell lines, namely, Caco-2 (CL-0050, Pricella, Wuhan, China), SW480 (CL-0223, Pricella, Wuhan, China), Lovo (CL-0144, Pricella, Wuhan, China), and HCT116 (CL-0096, Pricella, Wuhan, China), were cultured using Roswell Park Memorial Institute-1640 medium containing 10% fetal bovine serum (FBS; PM150110B, Pricella, Wuhan, China) in an incubator (5% carbon dioxide (CO2) and 37°C). All cells were identified by short-tandem repeat testing, and mycoplasma testing was conducted to prevent cell contamination.
Short hairpin (sh) RNA of TRAF4 (sh-TRAF4), TRAF4 overexpression (oe-TRAF4), PKM2 overexpression (oePKM2), and negative control (sh-NC and oe-NC) plasmids were obtained from GenePharma (GeneChem Co., Ltd, Shanghai, China). Cells (3 × 104 cells/well) were uniformly inoculated into 24-well plates. When the cell growth and fusion degree were approximately 70%, transfection was performed using Lipofectamine 2000 (11668-027, Invitrogen, Carlsbad, CA, USA). The cells were collected for experimental study after 48 h of transfection. In addition, wingless-type (Wnt)/β-catenin signal suppressor (XAV939; #3004, Sigma-Aldrich, St Louis, MO, USA) was added during rescue experiments to confirm whether the regulation of CRC cells’ malignant behavior and aerobic glycolysis by TRAF4 is related to the Wnt/β-catenin pathways. The cells were divided into control, sh-NC, oeNC, sh-TRAF4, oe-TRAF4, oe-PKM2, sh-TRAF4 + oe-NC, sh-TRAF4 + oe-PKM2, oe-TRAF4 + XAV939, and oePKM2 + XAV939 groups. The coding sequences of TRAF4 and PKM2 are shown in the supplementary material: shTRAF4: 5’-GCCCTGCACCTACTGCACTAA-3’ and sh-NC: 5’-TTCTCCGAACGTGTCACGT-3’.
Western blot
Cells were cleaved with a radioimmunoprecipitation assay lysate (R0010, Solarbio, Beijing, China), and cell protein concentration was measured using a bicinchoninic acid kit (P0010, Beyotime, Shanghai, China). The same amount of protein in each group was isolated through sodium dodecyl-sulfate polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride membrane (IPFL00010, Millipore, Billerica, MA, USA) through electric transfer. After being blocked, the primary antibodies PKM2 (1:1000, A20991, ABclonal, Wuhan, China), TRAF4 (1:1000, ab245666, Abcam, Waltham, MA, USA), glucose transporter type 1 (GLUT1; 1:1000, A11208, ABclonal, Wuhan, China), hexokinase 2 (HK2; 1:1000, A0994, ABclonal, Wuhan, China), lactate dehydrogenase A (LDHA; 1:1000, A1146, ABclonal, Wuhan, China), β-catenin (1:100, A11512, ABclonal, Wuhan, China), transcription factor Myc proto-oncogene (c-Myc) (1:1000, A17332, ABclonal, Wuhan, China), cyclin D1 (1:1000, A1301, ABclonal, Wuhan, China), and β-actin (1:1000, AC006, ABclonal, Wuhan, China) were added and reacted overnight at 4°C. After the reaction solution was incubated with the secondary antibody (1:2000, ab205718, Abcam, Waltham, MA, USA), the strips were exposed through enhanced chemiluminescence color development (E411-04, Vazyme, Nanjing, China). ImageJ software (1.43, NIH, Bethesda, MD, USA) was used in analyzing the gray values of protein bands, and β-actin was used as the internal reference.
Co-immunoprecipitation (Co-IP)
By searching for proteins that interact with TRAF4 through the protein-protein interaction database HitPredict (http://www.hitpredict.org/), we found that PKM2 can interact with TRAF4. TRAF4 and PKM2 interactions were detected through Co-IP. To precipitate the target protein, the antibodies of TRAF4 and PKM2 were incubated with the total protein successively, and the interacting proteins PKM2 and TRAF4 were detected separately. The cells were lysed and centrifuged for 20 min at 12,000 g at 4°C. TRAF4 (1:100, ab245666, Abcam, Waltham, MA, USA), PKM2 (1:100, A20991, ABclonal, Wuhan, China), or IgG antibody (1:100, #2729, Danvers, MA, USA; negative control) was added, and the cells were incubated at 4°C overnight with slow shaking. Protein A/G agarose beads (sc-2003, Santa Cruz, Dallas, TX, USA) pretreated with a buffer solution were added to the supernatant incubated with antibody for 2-4 h at 4℃ to make the antibody and protein A/G agarose beads conjugated. After the immunoprecipitation reaction, the agarose beads were centrifuged at 4°C with 12,000 g for 30 s. After the supernatant was removed, the agarose beads were washed 3 or 4 times with a cracking buffer. Finally, protein expression was analyzed with Western blot.
Real-time quantitative polymerase chain reaction
Total RNA was extracted according to the Trizol kit instructions (15596018CN, Invitrogen, Carlsbad, CA, USA). RNA purity and concentration were detected by an ultra-microspectrophotometer detector (NanoDrop 2000, Thermo Scientific, Wilmington, DE, USA). Total RNA (1 μg) was reversely transcribed into the first complementary deoxyribonucleic acid (cDNA) strand using a reverse transcription kit (RR037A, Takara, Beijing, China). The reverse transcription products were amplified with a synergetic binding reagent fluorescence quantitative polymerase chain reaction (PCR) kit (QR0100, Sigma, Saint Louis, MO, USA) on an ABI7500 fluorescence quantitative PCR amplification apparatus (7500, ABI, New York, NY, USA) for real-time quantitative polymerase chain reaction analysis. In the same sample, relative gene expression was assessed with the 2−ΔΔCt method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference.
Primers of TRAF4
Forward: 5’-CACAGGTGCCCTAAGCTGG-3’
Reverse: 5’-GGCTGAAGCACTCAAGGTTG-3’
Primers of GAPDH
Forward: 5’-TGCACCACCAACTGCTTAGC-3’
Reverse: 5’-GGCATGGACTGTGGTCATGAG-3’
Colony formation
Each group of cells was seeded into a six-well plate (1 × 103 cells/well) and cultured for 2 weeks. The culture medium was changed every 3-4 days, and cell morphology was observed. After cultivation, cells were fixed with paraformaldehyde (P0099, Beyotime, Shanghai, China), and stained with crystal violet (mlsw-1785, mlBio, Shanghai, China). After being washed and dried, the cells were observed and counted (more than 50 cells per clone).
5-ethynyl-2'-deoxyuridine (EdU)
Cell proliferation was detected with an EdU detection kit (C10310, RiboBio, Guangzhou, China). The transfected cells of each group were inoculated into 96-well plates (4 × 104 cells/well). After cell adhesion, EdU was added. Then, the cells were incubated for 2 h, and then, 4% paraformaldehyde was added to fix the cells. The cells were subjected to permeable treatment, and Apollo staining was performed for 30 min. After washing, DNA staining was performed with 4’-6-diamidino-2-phenylindole (C1005, Beyotime, Shanghai, China). Then, the excess staining solution was removed. Positive cells were photographed with an inverted fluorescence microscope (CKX53, Olympus, Tokyo, Japan).
Scratch healing assay
Cells (1 × 105) were inoculated into six-well plates and cultured conventionally. When the cells were 80% fused, lines were marked on the cell surface with a 10 μL gun tip. Distances between cells were measured after 24 h of culture. Scratch healing rate = (0 h scratch distance − 24 h scratch distance)/0 h scratch distance × 100%.
Transwell assay
The upper chamber of Transwell (G4740, Solarbio, Beijing, China) was coated with Matrigel matrix gel (356234, Solarbio, Beijing, China), 200 μL of cell suspension (1 × 105 cells/mL) was added to the upper chamber, and 800 μL culture medium containing 10% FBS was added to the lower chamber. After 24 h incubation, cells were fixed with paraformaldehyde and dyed with 0.1% crystal violet solution. After washing, non-invasive cells were wiped away with cotton swabs and transmembrane cells were observed with a microscope.
Glucose uptake and lactate production measurement
The transfected cells of each group were digested, centrifuged, and then suspended after the supernatant was discarded. The cells were inoculated into six-well plates at an adjusted density of 1 × 105 cells/well and cultured in an incubator at 37°C and 5% CO2. The cell culture supernatant of each group was collected after routine culture for 24 h. The concentrations of glucose and lactic acid in each group were measured strictly according to the protocols of the glucose assay reagent kit (abs580025, absin, Shanghai, China) and lactate assay kit (PM13313, PERFEMIKER, Shanghai, China). The corresponding reagents were added and incubated in sequence, and absorbance at 505 and 530 nm was detected using a multifunctional enzyme labeler (Synergy, BioTek, Burlington, VT, USA). Finally, the relative levels of glucose consumption and lactic acid generation were calculated.
Statistical analysis
GraphPad Prism statistical software (version 8.0, GraphPad Software Inc., San Diego, CA, USA) was used in analyzing data. Each experiment was repeated 3 times. Measurement data are expressed as mean ± standard deviation. Comparison between two groups was performed using t-test and one-way analysis of variance with Tukey’s post hoc test was used for multiple-group comparison. P < 0.05 was considered statistically significant.
RESULTS
TRAF4 expression in CRC cells
TRAF4 messenger ribonucleic acid (mRNA) and protein expression levels notably increased in the CRC (Caco-2 (P < 0.05, P < 0.001), SW480 (P < 0.001, P < 0.0001), Lovo (P < 0.0001), and HCT116 cells (P < 0.0001), and the Lovo cells had the highest TRAF4 expression level [Figures 1a-c]. Therefore, the Lovo cells were used as the research object in the following study.
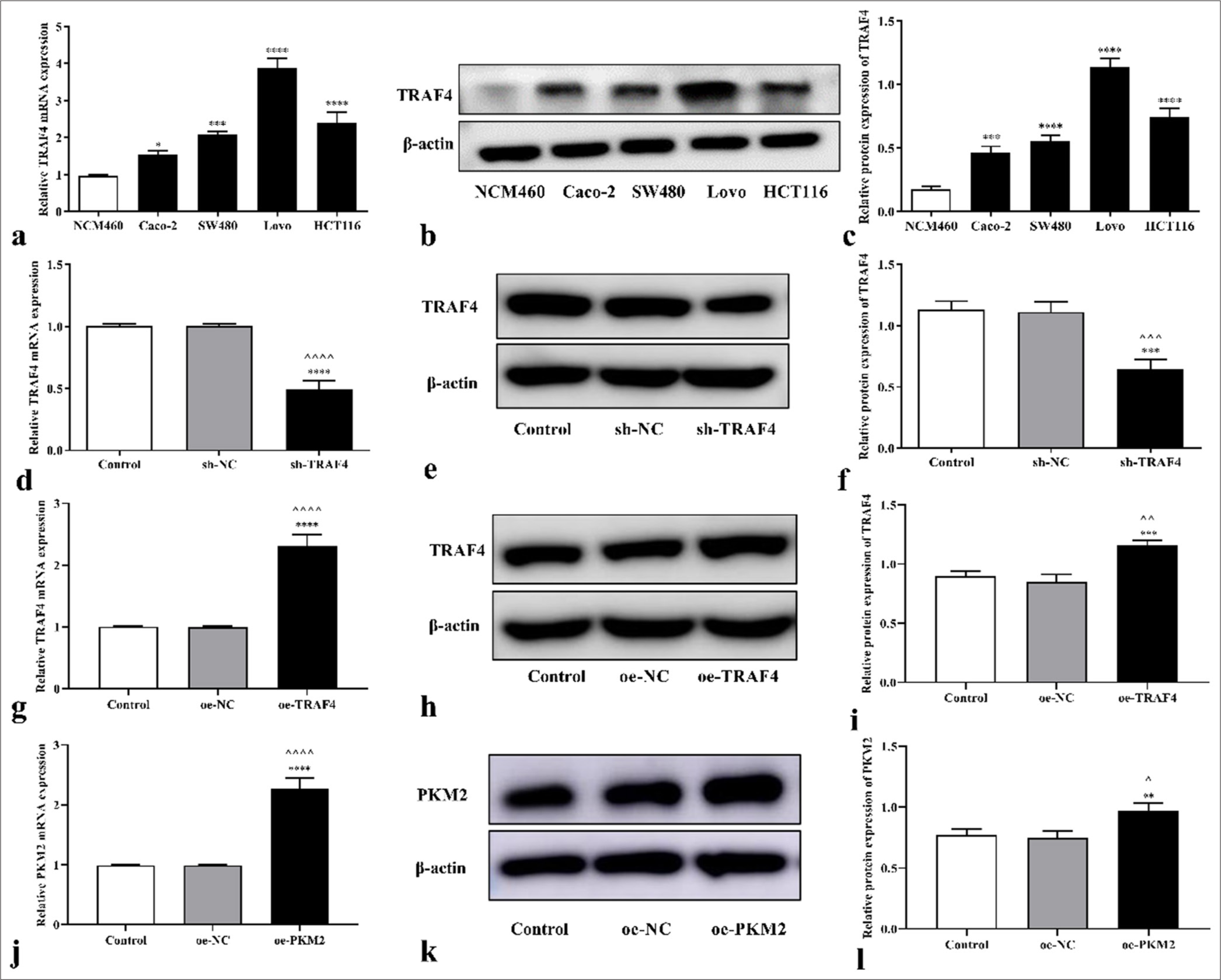
- Expression levels of TRAF4 in CRC cells. (a-c) TRAF4 mRNA and protein expression levels in CRC cells (n = 3). (d-l) TRAF4 or PKM2 expression was detected in CRC cells transfected with sh-TRAF4, oe-TRAF4, or oe-PKM2 (n = 3). ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, ^^^^P < 0.0001, versus the control group. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001, ✶✶✶✶P < 0.0001, versus the NCM460, sh-NC, or oe-NC group. TRAF4: Tumor necrosis factor receptor-associated factor 4, PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, NC: Negative control, sh: Short hairpin, oe: Overexpression.
TRAF4 interference treatment was performed on the Lovo cells, and how TRAF4 affects CRC progression was determined. After transfection with sh-TRAF4 and oeTRAF4, TRAF4 mRNA and protein expression levels in the cells decreased (P < 0.001, P < 0.0001) and increased (P < 0.01, P < 0.001, P < 0.0001) relative to those in the control, sh- NC, or oe-NC [Figures 1d-i]. Moreover, PKM2 mRNA (P < 0.0001) and protein (P < 0.05, P < 0.01) expression levels in the cells increased after oe-PKM2 transfection [Figures 1jl]. This result indicated that sh-TRAF4, oe-TRAF4, or oePKM2 were successfully transfected into the cells.
TRAF4 knockdown inhibited CRC cell malignant behavior and aerobic glycolysis
To clarify how TRAF4 affects CRC cells’ ability to proliferate, plate cloning and EdU cell proliferation experiments on CRC cells with differential expression of TRAF4 were conducted. TRAF4 knockdown inhibited cell colony-forming ability (P < 0.0001) [Figures 2a and b]. Further EdU cell proliferation experiments showed that TRAF4 knockdown inhibited cell proliferation ability (P < 0.0001) [Figures 2c and d]. TRAF4 knockdown also suppressed Lovo cell migration (P < 0.01) and invasion abilities (P < 0.0001) [Figures 3a-d].
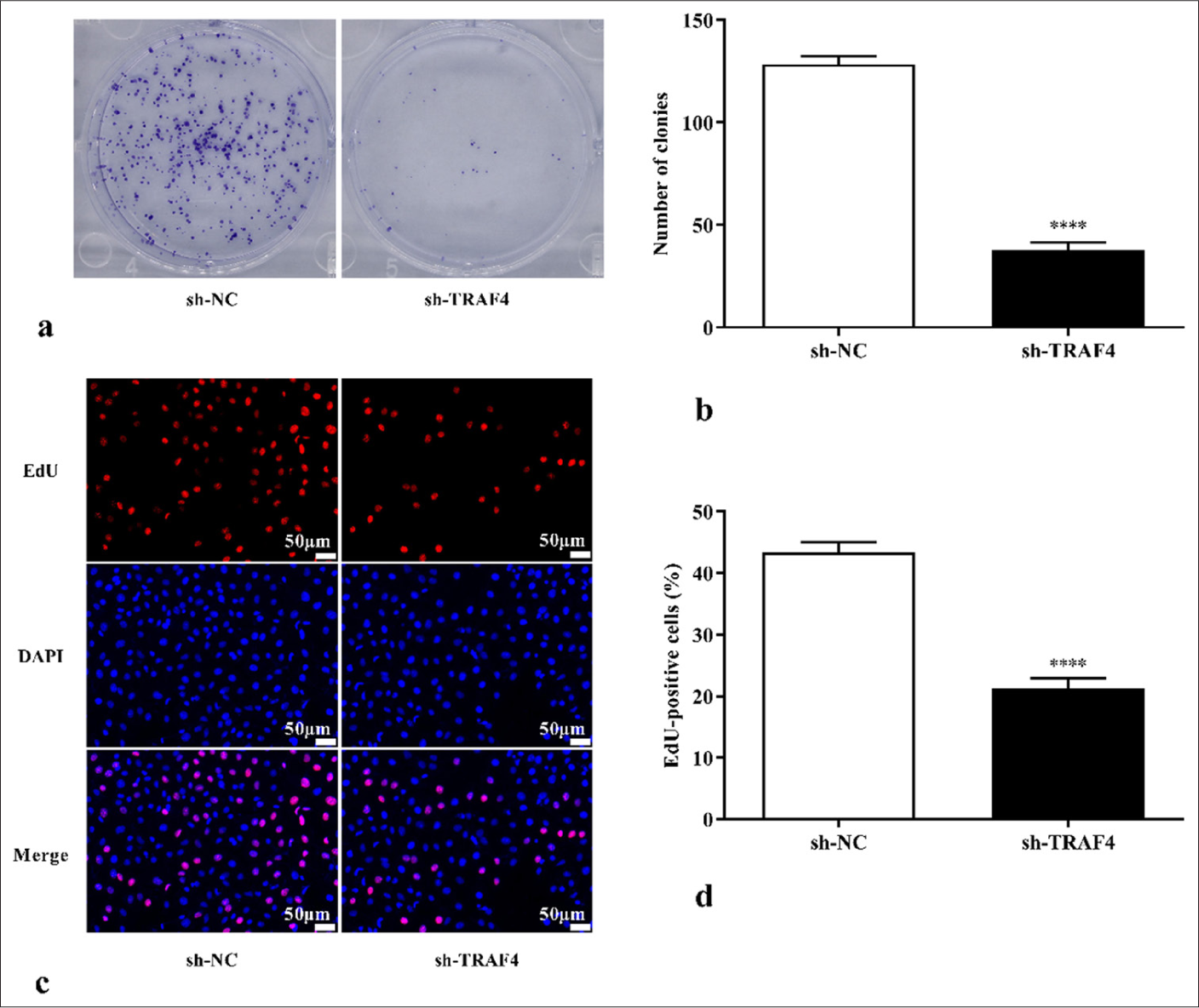
- TRAF4 knockdown inhibited CRC cell proliferation. (a-d) Colony formation and EdU assay, 200×, (scale bar: 50 μm) results showing that TRAF4 knockdown inhibited CRC cell proliferation (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. TRAF4: Tumor necrosis factor receptor-associated factor 4, EdU: 5-ethynyl-2’-deoxyuridine, DAPI: 4’-6-diamidino-2-phenylindole, NC: Negative control, sh: Short hairpin, CRC: Colorectal cancer.
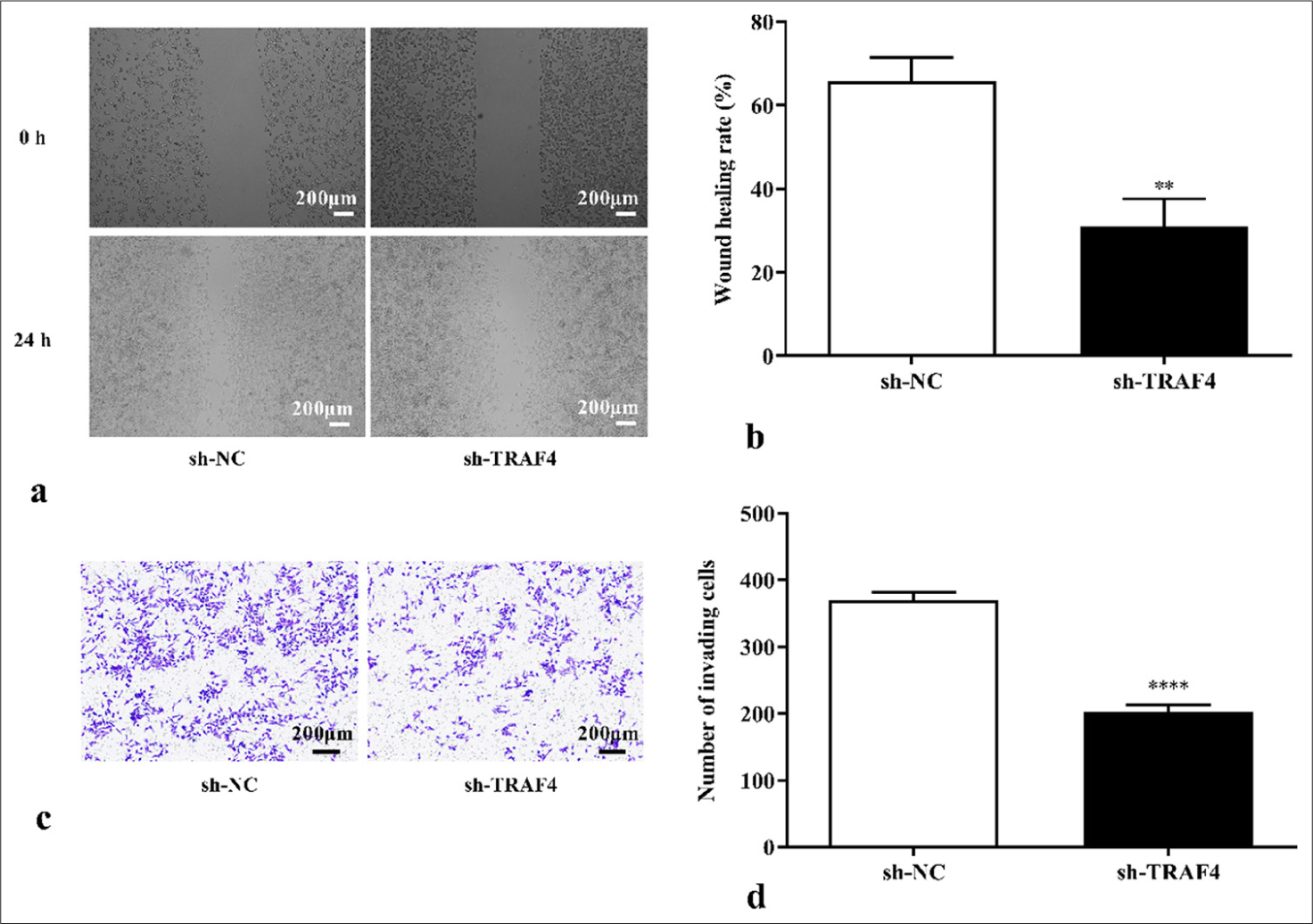
- TRAF4 knockdown inhibited CRC cell migration and invasion. (a-d) Cell migration and invasion abilities were measured using scratch healing, 40×, (scale bar: 200 μm) and Transwell assays, 40×, (scale bar: 200 μm) (n = 3). ✶✶P < 0.01, ✶✶✶✶P < 0.0001, versus sh-NC group. TRAF4: Tumor necrosis factor receptor-associated factor 4, CRC: Colorectal cancer, NC: Negative control, sh: Short hairpin.
Moreover, TRAF4 knockdown in Lovo cells reduced glucose uptake (P < 0.01) and lactate content (P < 0.01) [Figures 4a and b]. TRAF4 knockdown inhibited glycolysis-related expression levels of GLUT1 (P < 0.0001), PKM2 (P < 0.001), HK2 (P < 0.01), and LDHA (P < 0.0001) [Figures 4c and d]. These results suggested that TRAF4 knockdown inhibited malignant behavior and aerobic glycolysis of CRC cells.
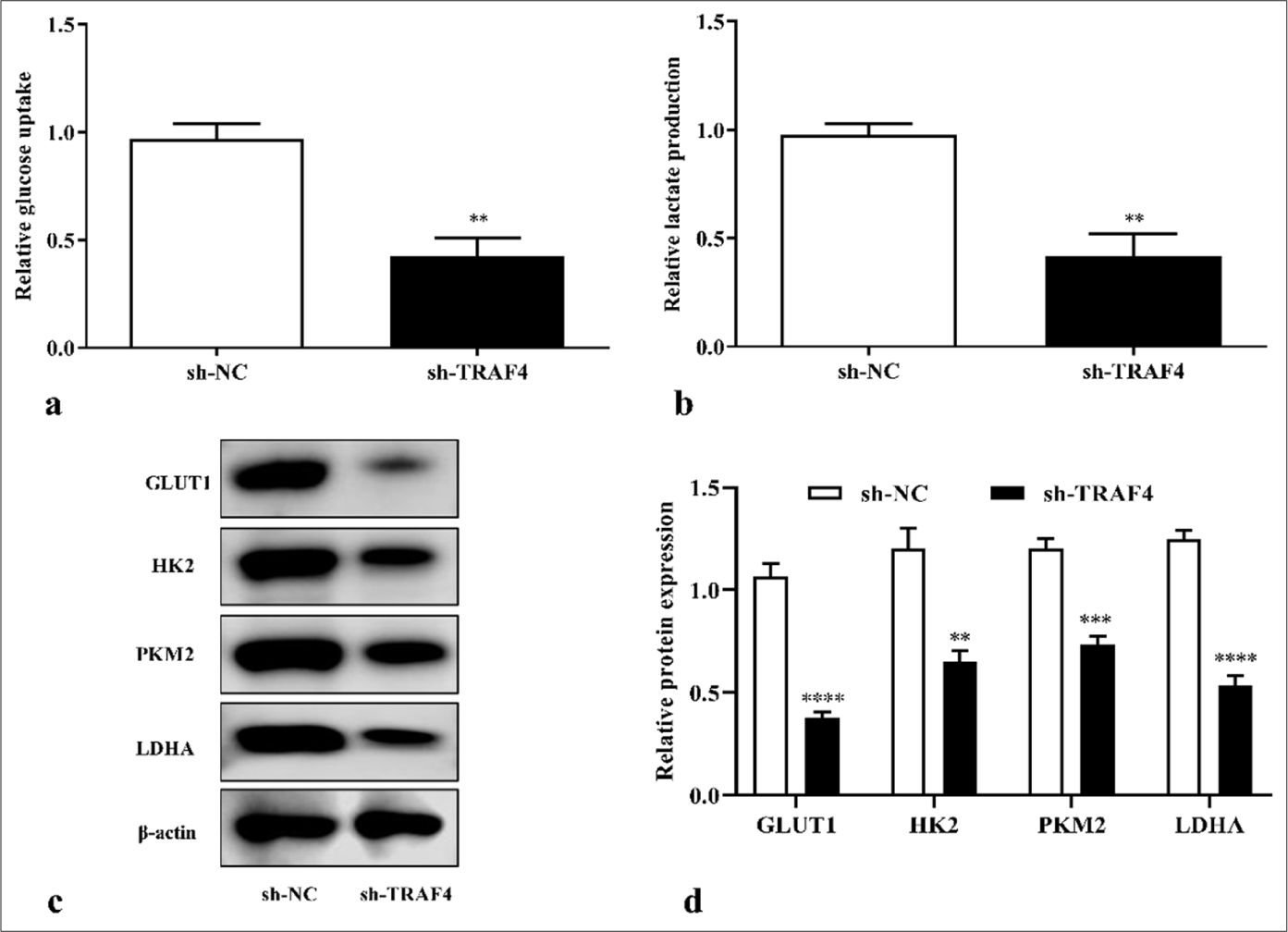
- TRAF4 knockdown inhibited aerobic glycolysis of CRC cells. (a-d) TRAF4 knockdown suppressed glucose uptake, lactate production, and glycolysis-related protein expression in CRC cells (n = 3). ✶✶P < 0.01, ✶✶✶P < 0.001, ✶✶✶✶P < 0.0001, versus sh-NC group. GLUT1: Glucose transporter type 1, HK2: Hexokinase 2, LDHA: Lactate dehydrogenase A, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, NC: Negative control, sh: Short hairpin.
TRAF4 can interact with PKM2
PKM2 interacted with TRAF4 in the Lovo cell [Figures 5a and b]. To clarify the regulatory relationship between TRAF4 and PKM2, TRAF4 overexpression or knockdown was performed in the Lovo cells. TRAF4 knockdown inhibited PKM2 protein expression (P < 0.0001), whereas TRAF4 overexpression promoted PKM2 protein expression (P < 0.001), indicating that TRAF4 positively regulated PKM2 expression [Figures 5c-f].
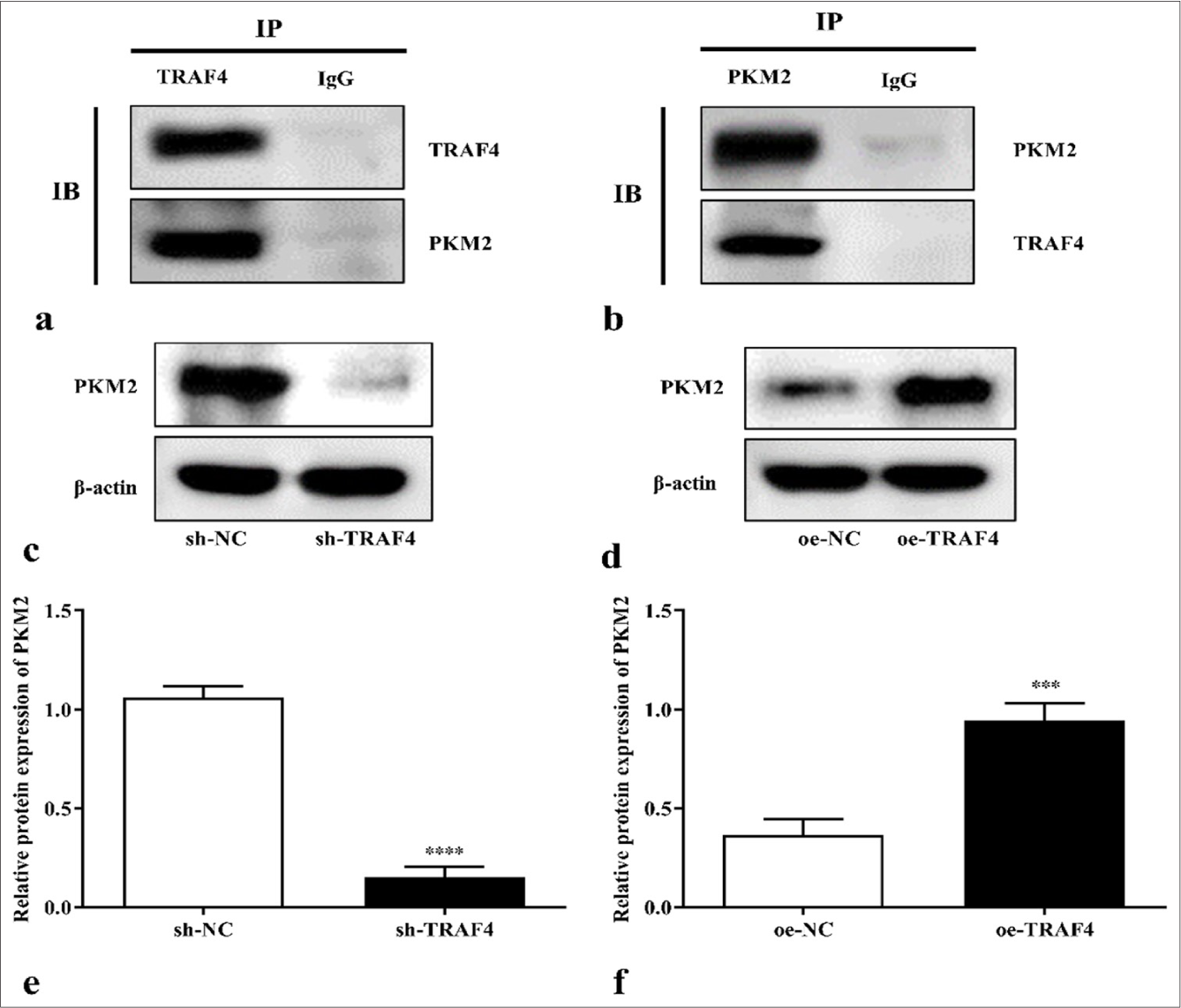
- TRAF4 can interact with PKM2. (a and b) Interactions of TRAF4 and PKM2 in CRC cells were analyzed by co-immunoprecipitation (n = 3). (c-f) PKM2 protein expression in CRC cells after sh-TRAF4 or oe-TRAF4 transfection (n = 3). ✶✶✶P < 0.001, ✶✶✶✶P < 0.0001, versus sh-NC or oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, NC: Negative control, sh: Short hairpin.
PKM2 overexpression restored the effects of TRAF4 knockdown on CRC cells’ malignant behavior and aerobic glycolysis
To further demonstrate that TRAF4 affects the Lovo cells’ malignant behavior and aerobic glycolysis through PKM2, oe- PKM2, and sh-TRAF4 were co-transfected into the Lovo cells. The co-transfection of oe-PKM2 and sh-TRAF4 can promote cell proliferation (P < 0.0001) [Figures 6a and b, 7a and b], migration (P < 0.01) [Figures 8a and b], invasion (P < 0.0001) [Figures 8c and d], glucose uptake (P < 0.05) [Figure 9a], and lactate production (P < 0.05) [Figure 9b] versus sh-TRAF4 and oe-NC co-transfection. Meanwhile, GLUT1 (P < 0.05), PKM2 (P < 0.01), HK2 (P < 0.0001), and LDHA (P < 0.0001) protein expression in the cells were promoted by sh-TRAF4 and oe-PKM2 co-transfection versus sh-TRAF4 and oe-NC co-transfection [Figures 9c and d]. In conclusion, PKM2 up-regulation reversed the effect of TRAF4 knockdown on cell malignant behavior and aerobic glycolysis.
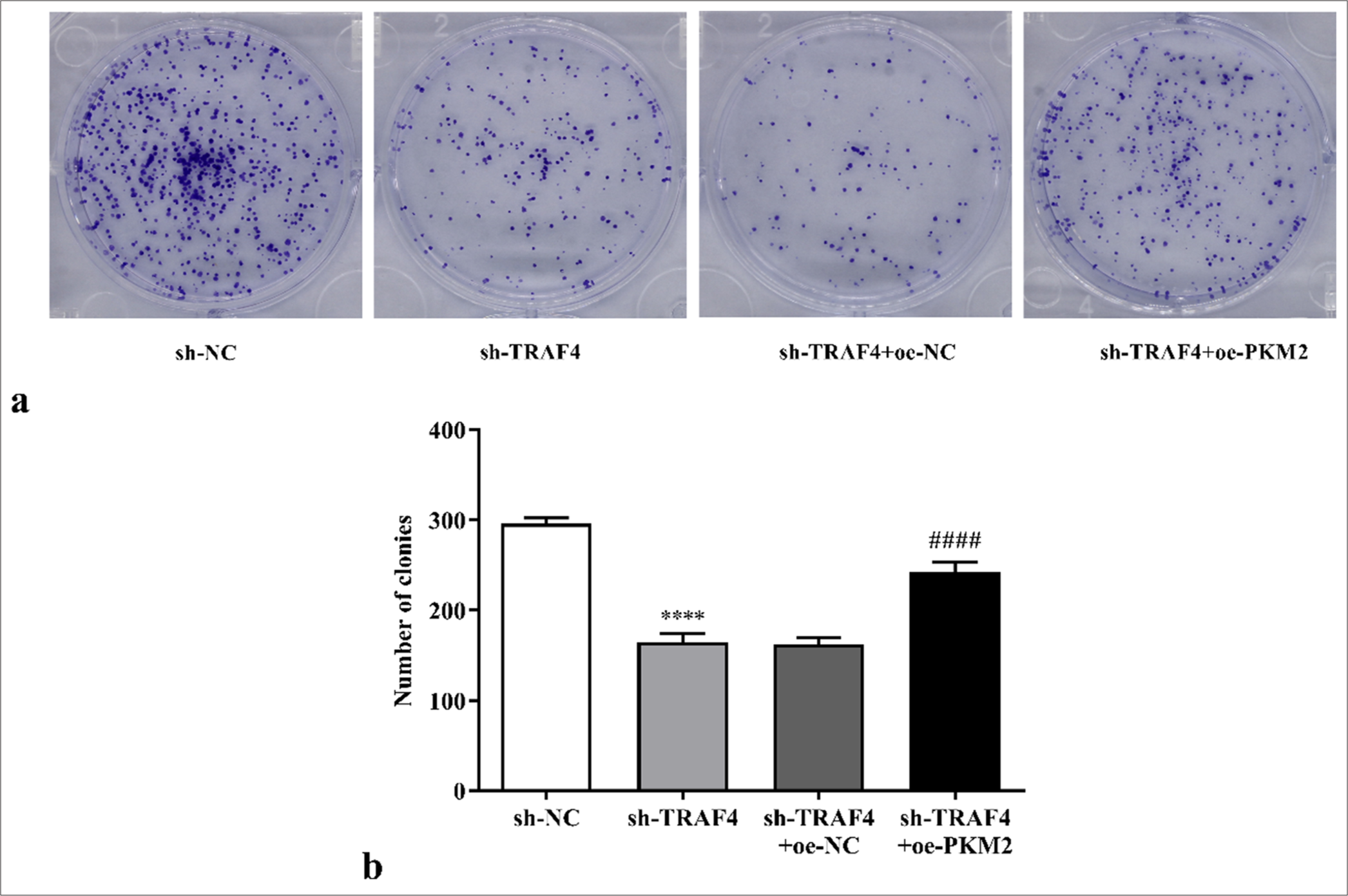
- Effects of sh-TRAF4 and oe-PKM2 co-transfection on CRC cell proliferation were detected through colony formation assay (a) clone formation images and (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. ####P < 0.0001, versus sh-TRAF4 + oeNC group. CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, PKM2: Pyruvate kinase muscle isoform 2, oe: Overexpression, NC: Negative control, sh: Short hairpin.
![Effects of sh-TRAF4 and oe-PKM2 co-transfection on CRC cell proliferation were detected with the EdU assay, 200×, (a) EdU images [scale bar: 50 μm], (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. ####P < 0.0001, versus sh-TRAF4 + oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, EdU: 5-ethynyl-2’-deoxyuridine, DAPI: 4’-6-diamidino-2-phenylindole, NC: Negative control, sh: Short hairpin.](/content/105/2025/22/1/img/Cytojournal-22-24-g007.png)
- Effects of sh-TRAF4 and oe-PKM2 co-transfection on CRC cell proliferation were detected with the EdU assay, 200×, (a) EdU images [scale bar: 50 μm], (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. ####P < 0.0001, versus sh-TRAF4 + oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, EdU: 5-ethynyl-2’-deoxyuridine, DAPI: 4’-6-diamidino-2-phenylindole, NC: Negative control, sh: Short hairpin.
![Changes in migration and invasion in the Lovo cells were detected after oe-PKM2 and sh-TRAF4 co-transfection (a) Cell scratch images, 40×, [scale bar: 200 μm], (b and c) statistical analysis of wound healing rate and number of invading cells, (d) Transwell invasion images, 40×, [scale bar: 200 μm]. (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. ##P < 0.01, ####P < 0.0001, versus sh-TRAF4 + oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, NC: Negative control, sh: Short hairpin.](/content/105/2025/22/1/img/Cytojournal-22-24-g008.png)
- Changes in migration and invasion in the Lovo cells were detected after oe-PKM2 and sh-TRAF4 co-transfection (a) Cell scratch images, 40×, [scale bar: 200 μm], (b and c) statistical analysis of wound healing rate and number of invading cells, (d) Transwell invasion images, 40×, [scale bar: 200 μm]. (n = 3). ✶✶✶✶P < 0.0001, versus sh-NC group. ##P < 0.01, ####P < 0.0001, versus sh-TRAF4 + oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, NC: Negative control, sh: Short hairpin.
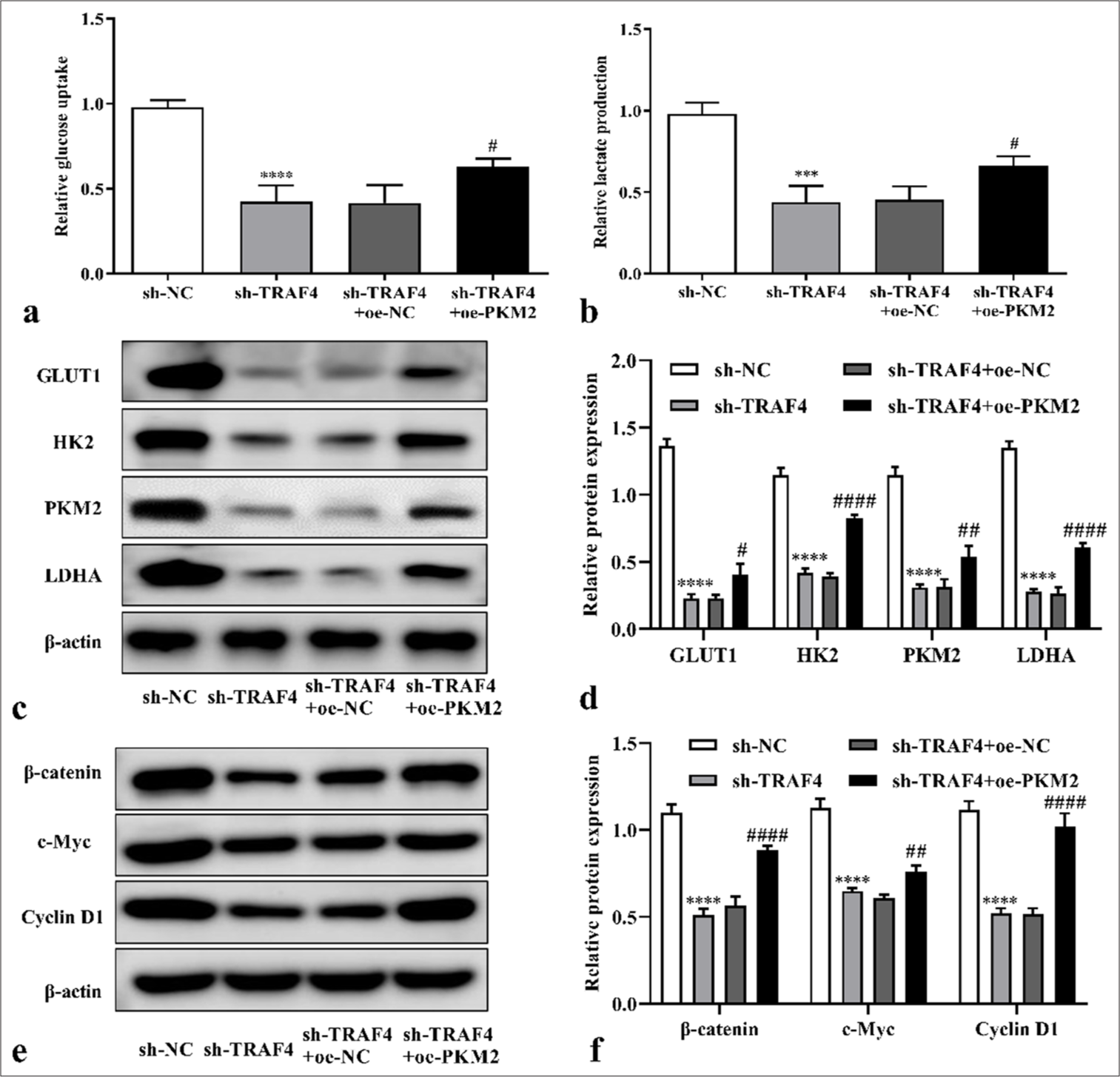
- PKM2 overexpression restored effect of TRAF4 knockdown on CRC cells’ aerobic glycolysis. (a and b) After PKM2 up-regulation in cells with TRAF4 knockdown, glucose uptake and lactate production were detected (n = 3). (c-f) Glycolytic-related and Wnt/β-catenin-related proteins expression in CRC cells after sh-TRAF4 + oe-PKM2 co-transfection were detected by Western blot (n = 3). ✶✶✶P < 0.001, ✶✶✶✶P < 0.0001, versus sh-NC group. #P < 0.05, ##P < 0.01, ####P < 0.0001, versus sh-TRAF4 + oe-NC group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, Wnt/β: Wingless-type/beta-catenin, oe: Overexpression, NC: Negative control, sh: Short hairpin.
Moreover, PKM2 regulates cancer development through the Wnt/β-catenin signaling pathway, which represents one regulator for glycolysis.[28] Thus, the level of Wnt/β-catenin pathway-related protein expression was detected, and whether TRAF4 affects that Wnt/β-catenin pathway was investigated. According to the results, β-catenin (P < 0.0001), c-Myc (P < 0.0001), and cyclin D1 (P < 0.0001) protein expression were inhibited by sh-TRAF4 [Figures 9e and f]. The co-transfection of oe-PKM2 and sh-TRAF4 can promote β-catenin (P < 0.0001), c-Myc (P < 0.01), and cyclin D1 (P < 0.0001) protein expression versus sh-TRAF4 and oe-NC co-transfection [Figures 9e and f].
TRAF4/PKM2/Wnt/β-catenin axis mediated cell malignant behavior and aerobic glycolysis
We speculated that TRAF4 may promote PKM2 expression and Wnt/β-catenin signaling activation and, thus, affected CRC cell malignant behavior and aerobic glycolysis. Therefore, a Wnt/β-catenin signal suppressor (XAV939) was introduced to the experiment. Here, oe-TRAF4 or oe-PKM2 promoted CRC cell proliferation (P < 0.0001) [Figures 10a and b, Figures 11a and b], migration (P < 0.001) [Figures 12a and b], invasion (P < 0.0001) [Figures 12c and d], glucose uptake (P < 0.0001) [Figure 13a], and lactate production (P < 0.0001) [Figure 13b] and the expression of GLUT1 (P < 0.0001), PKM2 (P < 0.0001), HK2 (P < 0.0001), and LDHA (P < 0.0001) proteins [Figures 13c and d]. The addition of XAV939 suppressed the promoting effects of oe-TRAF4 or oe-PKM2 on cell proliferation (P < 0.0001) [Figures 10a and b, Figures 11a and b], migration (P < 0.05, P < 0.01) [Figures 12a and b], invasion (P < 0.001, P < 0.0001) [Figures 12c and d], glucose uptake (P < 0.001) [Figure 13a], lactate production (P < 0.001, P < 0.01) [Figure 13b], and the expression of GLUT1 (P < 0.0001), PKM2 (P < 0.0001), HK2 (P < 0.0001), and LDHA (P < 0.0001) protein expression [Figures 13c and d].
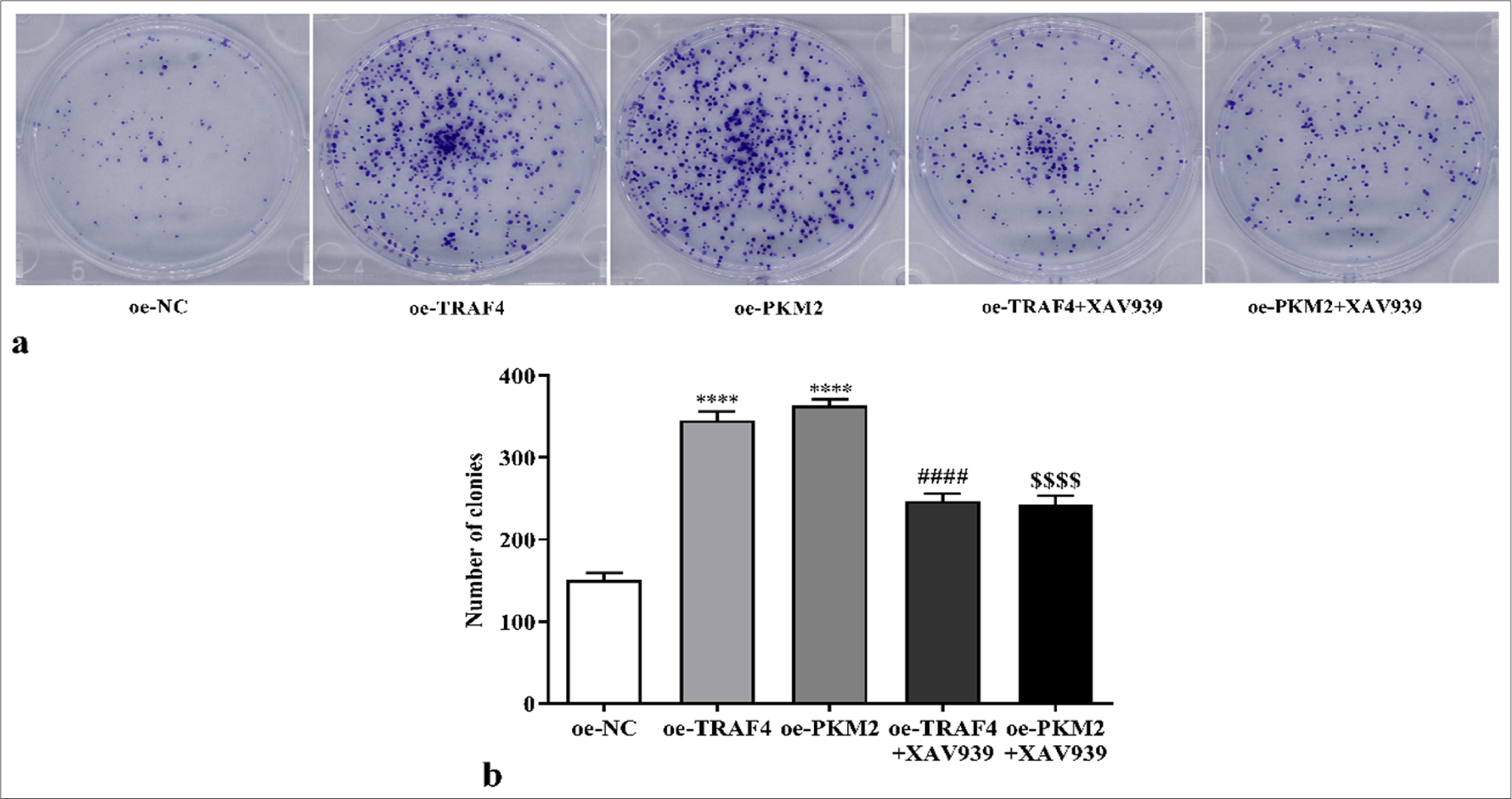
- Proliferation of CRC cells with oe-TRAF4, oe-PKM2, oe-TRAF4 and XAV939, or oe-PKM2 and XAV939 treatment was measured with colony formation assay (a) clone formation images, (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus oeNC group. ####P < 0.0001, versus oe-TRAF4 group. $$$$P < 0.0001, versus oe-PKM2 group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, NC: Negative control.
![Proliferation of CRC cells with oe-TRAF4, oe-PKM2, oe-TRAF4 and XAV939, or oe-PKM2 and XAV939 treatment was measured by EdU assays (a) EdU formation images, 200×, [scale bar: 50 μm], (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus oe-NC group. ####P < 0.0001, versus oe-TRAF4 group. $$$$P < 0.0001, versus oe-PKM2 group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, EdU: 5-ethynyl-2’-deoxyuridine, DAPI: 4’-6-diamidino-2-phenylindole, NC: Negative control.](/content/105/2025/22/1/img/Cytojournal-22-24-g011.png)
- Proliferation of CRC cells with oe-TRAF4, oe-PKM2, oe-TRAF4 and XAV939, or oe-PKM2 and XAV939 treatment was measured by EdU assays (a) EdU formation images, 200×, [scale bar: 50 μm], (b) statistical analysis. (n = 3). ✶✶✶✶P < 0.0001, versus oe-NC group. ####P < 0.0001, versus oe-TRAF4 group. $$$$P < 0.0001, versus oe-PKM2 group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, oe: Overexpression, EdU: 5-ethynyl-2’-deoxyuridine, DAPI: 4’-6-diamidino-2-phenylindole, NC: Negative control.
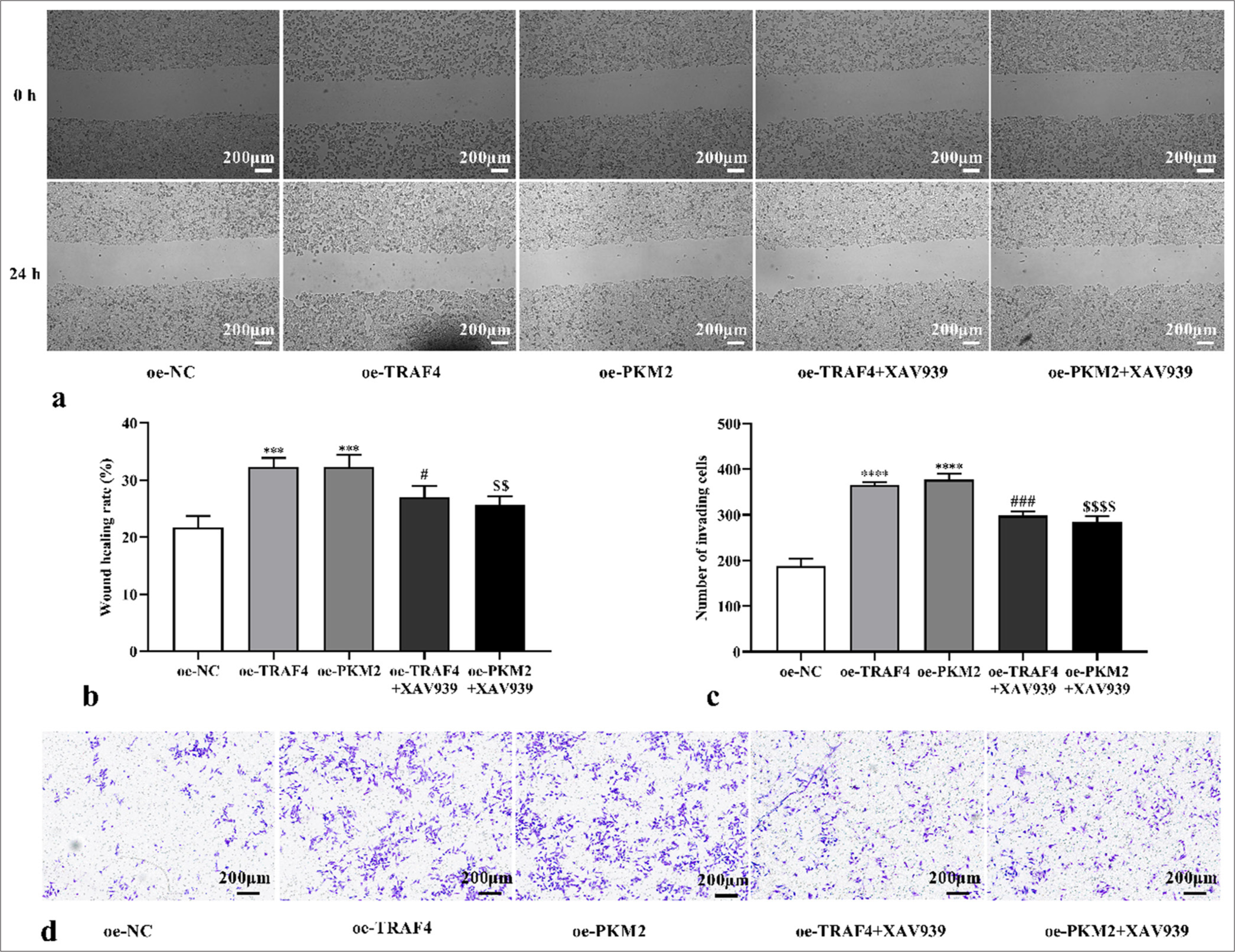
- TRAF4/PKM2/Wnt/β-catenin axis mediated cell migration and invasion. (a-d) CRC cell migration and invasion with oeTRAF4, oe-PKM2, oe-TRAF4 and XAV939, or oe-PKM2 and XAV939 treatment were assessed (scale bar: 200 μm) (n = 3). ✶✶✶P < 0.001, ✶✶✶✶P < 0.0001, versus oe-NC group. #P < 0.05, ###P < 0.001, versus oe-TRAF4 group. $$P < 0.01, $$$$P < 0.0001, versus oe-PKM2 group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, Wnt/β: Wingless-type/beta-catenin, oe: Overexpression, NC: Negative control.
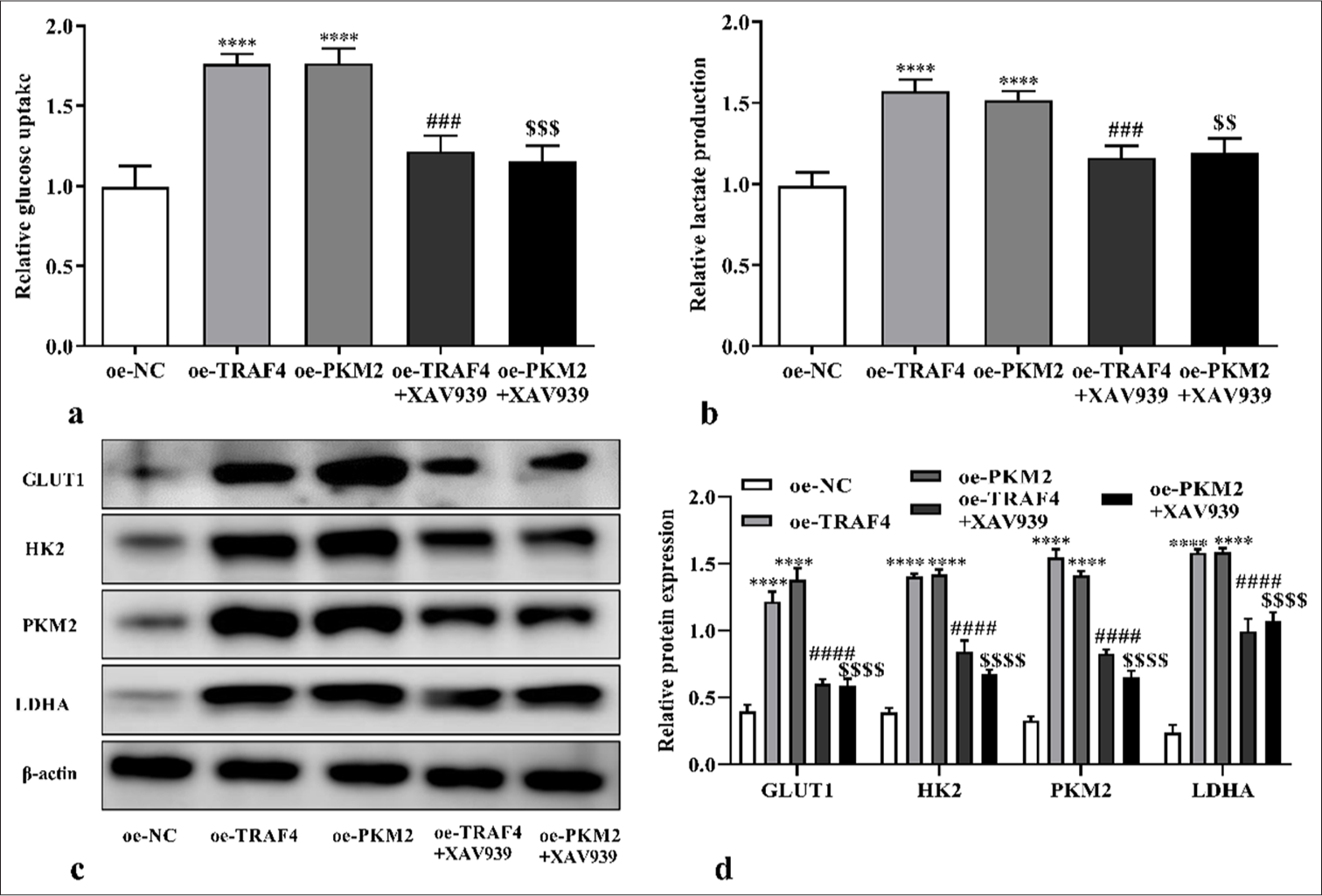
- TRAF4/PKM2/Wnt/β-catenin axis mediated aerobic glycolysis. (a-d) Glucose uptake, lactate production, and glycolytic-related proteins levels of CRC cells with oe-TRAF4, oe-PKM2, oeTRAF4 and XAV939, or oe-PKM2 and XAV939 treatments were assessed (n = 3). ✶✶✶✶P < 0.0001, versus oe-NC group. ###P < 0.001, ####P < 0.0001, versus oe-TRAF4 group. $$P < 0.01, $$$P < 0.001, $$$$P < 0.0001, versus oe-PKM2 group. PKM2: Pyruvate kinase muscle isoform 2, CRC: Colorectal cancer, TRAF4: Tumor necrosis factor receptor-associated factor 4, Wnt/β: Wingless-type/beta-catenin, oe: Overexpression.
DISCUSSION
CRC occurrence involves gene mutation and heredity, physical, chemical, and biological factors and is related to the living habits and nutritional levels of patients.[29] Finding novel molecular targets for inhibiting tumor occurrence and development is vital for improving the prognoses of patients with CRC. To maintain uncontrolled rapid proliferation, tumor cells need a considerable amount of energy and nutrients.[30,31] Aerobic glycolysis is crucial for the proliferation of various malignant tumor cells and may be a universal mechanism linking energy metabolism and the proliferative phenotype of tumor cells.[32] Thus, exploring the aerobic glycolysis mechanism in CRC may provide a novel approach for CRC treatment.
TRAF4 involves the occurrence of human malignant tumors and the abnormal metabolism of tumor cells.[13] Here, TRAF4 knockdown inhibited CRC cells’ malignant behavior, consistent with previous findings.[20-22] In addition, TRAF4 regulates glycolysis in lung cancer cells and promotes tumor progression.[33] Interestingly, we found that knocking down TRAF4 inhibits glycolysis in CRC, and PKM2 can interact with TRAF4. TRAF4 promoted PKM2 expression in CRC cells, and PKM2 up-regulation offset the effects of TRAF4 knockdown on cell malignant behavior and aerobic glycolysis. These results suggested that TRAF4 mediates tumor cells’ malignant behavior and aerobic glycolysis in CRC by regulating PKM2. Tribbles homolog 2 promotes lung cancer progression and glycolysis by interacting with PKM2 through phosphorylation.[34] In CRC, PKM2 stability and nuclear translocation are promoted by aspartateglutamate-x-aspartate/histidine-box polypeptide 39B.[35] Moreover, acute myeloid leukemia progression is inhibited by the Josephin domain containing 2 blocking PKM2 nuclear localization.[36] Meanwhile, we found a direct interaction between TRAF4 and PKM2 and observed that TRAF4 promoted the activation of PKM2. Then, we explored how TRAF4 regulates PKM2. To date, no report on the regulation of PKM2 by TRAF4 has been conducted, and further experiments are needed to explore its detailed molecular mechanism in the future.
The abnormal activation of the Wnt/β-catenin pathway is associated with the development of malignancies,[37] and the activation and shutdown of the Wnt/β-catenin pathway is related to the regulation of cell proliferation, tumor cell glycolysis, cancer initiation, development, deterioration, and metastasis.[38,39] In CRC, the TRAF4/Wnt/β-catenin pathway promotes growth and invasion.[22] Here, TRAF4 knockdown inhibited PKM2, cyclin D1, c-Myc, and β-catenin protein expression, but the levels were restored by PKM2 overexpression plasmid co-transfection. As expected, XAV939 reversed the effect of TRAF4 or PKM2 overexpression on CRC cells’ malignant behavior and aerobic glycolysis. Due to the limitations of funding, equipment, and time, this study has some deficiencies that need to be supplemented and improved in a follow-up study. In this study, TRAF4 was only down-regulated in the Lovo cells. This result needs to be further improved in more than one cell line. In the measurement of aerobic glycolysis function, only glucose uptake and lactate production were measured.
In current research on cellular glycolysis function, glycolysis efficiency can be measured in real-time using cellular acidification rate detection, mass spectrometry, and liquid phase analysis systems. This approach needs to be further improved. Moreover, whether the results of this study can be verified in animals remains to be further investigated in animal experiments. Most studies have their own limitations and are not perfect, and including all the experiments in a single study is impossible. Despite these limitations, this study has made some important discoveries about the role of TRAF4 in CRC. This study first proposed the relationship between TRAF4 and PKM2 in CRC through in vitro cell experiments and found an interaction between them. Next, we further explored the role of TRAF4 interaction with PKM2 in CRC development. In addition, TRAF4 regulated Wnt/β-catenin signaling pathway, and the response experiment found that PKM2 overexpression after TRAF4 knockdown promoted β-catenin, c-Myc, and cyclin D1 expression. Moreover, XAV939 addition partially inhibited the CRC cells’ malignant behavior and glycolysis induced by TRAF4 or PKM2 overexpression. These results demonstrated that TRAF4 promotes CRC progression through PKM2 and activates the Wnt/β-catenin signaling. This provides a basis for enriching the mechanism network of TRAF4-mediated glucose metabolism. Aerobic glycolysis is closely related to drug resistance, immune escape, and metastasis of tumor cells.[10] The effects of TRAF4 on drug resistance and immune microenvironment after inhibiting the aerobic glycolysis of CRC cells will be further studied in the future to provide certain theoretical basis and novel ideas for clinical breast cancer treatment and novel drug development.
SUMMARY
TRAF4 and PKM2 regulated CRC cell malignant behavior and aerobic glycolysis. TRAF4 and PKM2 proteins bind and promote CRC cell malignant behavior and aerobic glycolysis by Wnt/β-catenin signaling pathway. However, some common mediating factors or other regulatory modes between TRAF4 and PKM2 may exist. Therefore, the regulatory mechanism of TRAF4 should be further explored. The relationship between TRAF4 and the clinicopathologic features of patients with CRC has not been clearly defined and has not been further verified by animal experiments. In the future, it will be further explored and improved to provide a novel target for the targeted therapy of CRC.
AVAILABILITY OF DATA AND MATERIALS
The dataset that supports the results and findings of this research is available from the corresponding author on reasonable request.
ABBREVIATIONS
cDNA: Complementary deoxyribonucleic acid
c-Myc: Transcription factor Myc proto-oncogene
CO2: Carbon dioxide
Co-IP: Co-immunoprecipitation
CRC: Colorectal cancer
DAPI: 4’-6-diamidino-2-phenylindole
EdU: 5-ethynyl-2’-deoxyuridine
FBS: Fetal bovine serum
GAPDH-Glyceraldehyde-3-phosphate dehydrogenase
GLUT1: Glucose transporter type 1
HK2: Hexokinase 2
LDHA: Lactate dehydrogenase A
mRNA: Messenger ribonucleic acid
NC: Negative control
oe: Overexpression
PCR: Polymerase chain reaction
PKM2: Pyruvate kinase muscle isoform 2
sh-RNA: Short hairpin ribonucleic acid
TRAF4: Tumor necrosis factor receptor-associated factor 4
Wnt/β: Wingless-type/beta-catenin
ACKNOWLEDGMENTS
Not applicable.
AUTHOR CONTRIBUTIONS
YYM and TML: Designed the research study; TML, SHZ, JWS, and YYM: Performed the research; SHZ, JWS, and YYM: Collected and analyzed the data; YYM and TML: Drafting the manuscript; and all authors have been involved in revising it critically for important intellectual content. All authors give final approval of the version to be published. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Ethical approval and consent to participate is not required as this study does not involve animal or human experiments.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: Not applicable.
References
- Ferroptosis: Reviewing CRC with the third eye. J Inflamm Res. 2022;15:6801-12.
- [CrossRef] [PubMed] [Google Scholar]
- Dichotomous colorectal cancer behaviour. Crit Rev Oncol Hematol. 2023;189:104067.
- [CrossRef] [PubMed] [Google Scholar]
- Total saponins from Rhizoma Panacis Majoris inhibit proliferation, induce cell cycle arrest and apoptosis and influence MAPK signalling pathways on the colorectal cancer cell. Mol Med Rep. 2021;24:542.
- [CrossRef] [PubMed] [Google Scholar]
- Recent approaches on molecular markers, treatment and novel drug delivery system used for the management of colorectal cancer: A comprehensive review. Curr Pharm Biotechnol. 2024;25:1969-85.
- [CrossRef] [PubMed] [Google Scholar]
- The mechanism of miR-103a-3p regulating PFK-2 on proliferation and glycolysis of colorectal cancer cells. Chin J Gerontol. 2024;44:454-9.
- [Google Scholar]
- Subcellular localization of nucleolar protein 14 and its proliferative function mediated by miR-17-5p and E2F4 in pancreatic cancer. Aging (Albany NY). 2023;15:7308-23.
- [CrossRef] [PubMed] [Google Scholar]
- Flexibility in metabolism bestows tenacious viability on cancer. Life Sci. 2018;208:20-5.
- [CrossRef] [PubMed] [Google Scholar]
- ALDOB plays a tumor-suppressive role by inhibiting AKT activation in gastric cancer. J Cancer. 2023;14:2255-62.
- [CrossRef] [PubMed] [Google Scholar]
- GATA3/MiR-199a-3p suppresses glycometabolism in breast cancer by inhibiting PFKFB3 expression. J Biol Regul Homeost Agents. 2024;38:223-31.
- [CrossRef] [Google Scholar]
- Tumor glycolysis, an essential sweet tooth of tumor cells. Semin Cancer Biol. 2022;86:1216-30.
- [CrossRef] [PubMed] [Google Scholar]
- TRIM28 regulates hepatocellular carcinoma progression and aerobic glycolysis through interacting with PFKFB3. J Biol Regul Homeost Agents. 2023;37:951-9.
- [CrossRef] [Google Scholar]
- Tumor metabolism destruction via metformin-based glycolysis inhibition and glucose oxidase-mediated glucose deprivation for enhanced cancer therapy. Acta Biomater. 2022;145:222-34.
- [CrossRef] [PubMed] [Google Scholar]
- TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2022;26:2652-66.
- [CrossRef] [PubMed] [Google Scholar]
- Advacements in investigating the role of TRAF4 in facilitating tumorigenesis and progression. Chin J Clin Oncol. 2023;50:531-7.
- [Google Scholar]
- High expression of TRAF4 predicts poor prognosis in tamoxifen-treated breast cancer and promotes tamoxifen resistance. Anticancer Drugs. 2020;31:558-66.
- [CrossRef] [PubMed] [Google Scholar]
- Knockdown of TRAF4 expression suppresses osteosarcoma cell growth in vitro and in vivo. Int J Mol Med. 2014;34:1655-60.
- [CrossRef] [PubMed] [Google Scholar]
- TRAF4 promotes lung cancer development by activating tyrosine kinase of EGFR. Chin J Oncol. 2024;46:968-78.
- [Google Scholar]
- TRAF4 enhances oral squamous cell carcinoma cell growth, invasion and migration by Wnt-β-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8:11837.
- [Google Scholar]
- TRAF4 promotes the malignant progression of high-grade serous ovarian cancer by activating YAP pathway. Biochem Biophys Res Commun. 2022;627:68-75.
- [CrossRef] [PubMed] [Google Scholar]
- Ubiquitination of the DNA-damage checkpoint kinase CHK1 by TRAF4 is required for CHK1 activation. J Hematol Oncol. 2020;13:40.
- [CrossRef] [PubMed] [Google Scholar]
- TRAF4-mediated ubiquitination-dependent activation of JNK/Bcl-xL drives radioresistance. Cell Death Dis. 2023;14:102.
- [CrossRef] [PubMed] [Google Scholar]
- TRAF4 promotes the growth and invasion of colon cancer through the Wnt/β-catenin pathway. Int J Clin Exp Pathol. 2015;8:1419-26.
- [Google Scholar]
- Mechanism of PKM2 affecting cancer immunity and metabolism in Tumor Microenvironment. J Cancer. 2021;12:3566-74.
- [CrossRef] [PubMed] [Google Scholar]
- PKM2 promotes neutrophil activation and cerebral thromboinflammation: Therapeutic implications for ischemic stroke. Blood. 2022;139:1234-45.
- [CrossRef] [PubMed] [Google Scholar]
- PKM2, a potential target for regulating cancer. Gene. 2018;668:48-53.
- [CrossRef] [PubMed] [Google Scholar]
- Pyruvate kinase M2 (PKM2) in cancer and cancer therapeutics. Cancer Lett. 2021;503:240-8.
- [CrossRef] [PubMed] [Google Scholar]
- Research progress of pyruvate kinase M2 in colorectal cancer. J Gannan Med Univ. 2023;43:10-6.
- [Google Scholar]
- Knockdown of PKM2 suppresses tumor progression in human cervical cancer by modulating epithelial-mesenchymal transition via Wnt/β-catenin signaling. Cancer Manag Res. 2018;10:4191-202.
- [CrossRef] [PubMed] [Google Scholar]
- Research progress on the role of CMTM4 in colorectal cancer. Zhejiang Med J. 2024;46:95-7.
- [Google Scholar]
- Cancer metabolism: Looking forward. Nat Rev Cancer. 2021;21:669-80.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting glucose metabolism to suppress cancer progression: Prospective of anti-glycolytic cancer therapy. Pharmacol Res. 2019;150:104511.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells. 2021;10:1056.
- [CrossRef] [PubMed] [Google Scholar]
- TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938-50.
- [CrossRef] [PubMed] [Google Scholar]
- Oncogenic TRIB2 interacts with and regulates PKM2 to promote aerobic glycolysis and lung cancer cell procession. Cell Death Dis. 2022;8:306.
- [CrossRef] [PubMed] [Google Scholar]
- DDX39B drives colorectal cancer progression by promoting the stability and nuclear translocation of PKM2. Signal Transduct Target Ther. 2022;7:275.
- [CrossRef] [PubMed] [Google Scholar]
- JOSD2 regulates PKM2 nuclear translocation and reduces acute myeloid leukemia progression. Exp Hematol Oncol. 2022;11:42.
- [CrossRef] [PubMed] [Google Scholar]
- Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther. 2021;6:307.
- [CrossRef] [PubMed] [Google Scholar]
- Advances in targeting the WNT/β-catenin signaling pathway in cancer. Drug Discov Today. 2022;27:82-101.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165.
- [CrossRef] [PubMed] [Google Scholar]








