Translate this page into:
Investigating the role of exosomal microRNA-5703 in modulating tumor-associated endothelial cells in lung cancer

*Corresponding Author: Zhenfa Zhang, Department of Lung Cancer Surgery, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, Tianjin, China. zhangzhenfa@tmu.edu.cn
-
Received: ,
Accepted: ,
How to cite this article: Wen B, Tao R, Liu Y, Zhang Z. Investigating the role of exosomal microRNA-5703 in modulating tumor-associated endothelial cells in lung cancer. CytoJournal 2024;21:77. doi: 10.25259/Cytojournal_99_2024
Abstract
Objective:
Lung cancer, as a prevalent malignancy, continues to be a considerable clinical challenge. This study aimed to elucidate the role of microRNA-5703 (miR-5703) in lung cancer progression and to assess the effect of exosomal miR-5703 on tumor-associated endothelial cells (TAECs).
Material and Methods:
We analyzed Gene Expression Omnibus datasets and performed quantitative real-time polymerase chain reaction to determine miR-5703 expression levels in lung cancer tissues. Exosomes derived from lung cancer cells were identified, and the effects of miR-5703 inhibitors or mimics on malignant biological behavior were evaluated in the lung cancer cells. Moreover, to understand these effects on TAECs, we assessed angiogenesis, endothelial–mesenchymal transition (EndMT), and barrier function after treatment with miR- 5703 inhibitors or the exosome-assimilated inhibitor cytochalasin D. Tumor-bearing mouse models were used in validating the tumor-promoting effects of exosomes derived from lung cancer cells, and the markers of angiogenesis, EndMT, and barrier function were examined.
Results:
Our results showed that miR-5703 was up-regulated in the lung cancer cells and patient-derived exosomes. miR-5703 facilitated cell growth, migration, invasion, in LC cells, and impaired the barrier function, which promoted angiogenesis and EndMT of TAECs by carrying in exosomes through targeting inhibitor of growth family member 4 (ING4) was identified as target of miR-5703 (P < 0.05). In vivo, the tumor-promoting effects of lung cancer cell–derived exosomes were rescued by miR-5703 inhibitors, leading to the up-regulation of ING4 expression and reduction in vascular distribution in the tumor tissues (P < 0.05).
Conclusion:
miR-5703 operates as an oncogenic factor in lung cancer. After being taken up by TAECs, exosomal miR-5703 promotes angiogenesis, EndMT, and barrier damage by targeting ING4. Hence, miR-5703 is a potential target in the lung cancer microenvironment.
Keywords
lung cancer
exosome
tumor-associated endothelial cell
MicroRNA
INTRODUCTION
In the past decades, lung cancer has been the most frequently diagnosed cancer and the primary cause of cancer-related mortality globally.[1] It is initially asymptomatic and typically discovered at advanced stages[2] and is often associated with risk factors, such as long-term smoking; exposure to second-hand smoke, environmental toxins, and ionizing radiation; human immunodeficiency virus infection; alcohol consumption; and genetic factors.[3] Lung cancer treatments include surgical removal, chemotherapy, radiation therapy, and therapy targeting specific cancer characteristics.[4] Treatment selection is influenced by patients’ conditions, tumor type and stage, and overall health.[5] However, the 5-year survival rate of lung cancer remains poor.[6] Metastasis and resistance to treatment present obstacles to lung cancer management.[7] Thus, elucidating the pathogenesis and progression of lung cancer is imperative.
Tumor-associated endothelial cells (TAECs) are crucial for tumor development and metastasis, possessing the following unique characteristics: (1) Enhanced angiogenic capacity: Stimulated by hypoxia and inflammation within the tumor microenvironment, TAECs overexpress angiogenic factors, and promoting neovascularization. TAECs enhance this capability through autocrine and paracrine mechanisms.[8] (2) Impaired barrier function: The reduced expression or functionality of junction proteins in TAECs diminishes vascular integrity and permeability.[9] (3) Endothelial–mesenchymal transition (EndMT) characteristics: TAECs can undergo EndMT, gaining migratory and invasive capabilities, facilitating tumor vascular remodeling and metastasis, which is characterized by a decrease in endothelial cell markers and an increase in mesenchymal cell markers.[10] These traits collectively favor tumor growth and metastasis.[11]
Exosomes (30–150 nm) are small extracellular vesicles released by various cells, including normal and cancer cells, into the extracellular environment.[12] They are among the components of the body’s intercellular communication system and deliver multiple functional biomolecules, including proteins, lipids, and nucleic acids.[13] Most cells release exosomes, whereas tumor cells may produce more exosomes to promote their own survival and invasion under hypoxic conditions.[14] Some researchers have highlighted the role of hypoxic tumor cell-derived exosomes in promoting angiogenesis.[15-17]
A microRNA (miRNA) operates by specifically attaching to a messenger RNA (mRNA), resulting in the degradation of the mRNA or suppression of its translation into a protein.[18] Cancer-derived exosomal miRNAs are involved in cancer metastasis and progression. For example, exosomal microRNA-5703 (miR-5703) has been demonstrated as an onco-miRNA in pancreatic cancer.[19] However, the function of exosomal miRNAs has not been fully characterized in lung cancer angiogenesis, tumor growth, and metastasis.
In this study, we investigated the expression pattern of miR-5703 in lung cancer and determine whether lung cancer cell-derived exosomes can induce the angiogenesis, tumorigenesis, and EndMT of TAECs. We showed that exosomal miR-5703 down-regulated the expression of inhibitor of growth family member 4 (ING4) in endothelial cells and thereby promoted lung cancer progression and angiogenesis. These results strongly indicate that exosomal miR-5703 is a prognostic indicator for tumor progression and a therapeutic target.
MATERIAL AND METHODS
Sampling from patients
A cohort of patients with lung cancer was meticulously selected using stringent criteria. Each patient underwent an exhaustive pre-operative clinical evaluation encompassing chest radiography, computed tomography, and pulmonary function test for the accurate diagnosis of lung carcinoma and precise delineation of tumor dimensions and locations. Apart from diagnostic modalities, an in-depth collection of patients’ clinical attributes was undertaken. The data encompassed age, gender, metastatic status, tumor dimension, and clinical stage. On the confirmation of lung cancer diagnosis, patients were apprised in detail regarding the study’s objectives and methodologies. The study was conducted in compliance with the Declaration of Helsinki. Informed consent was obtained from all study participants, and ethics approval was obtained from the Ethics Committee of the Second People’s Hospital of Yibin (no. 2023-162-01). During surgical intervention, paired tissue specimens, cancer tissues (n = 20), and adjacent normal tissues (n = 20) were harvested. Blood samples were drawn from each patient or healthy individual (n = 20), and serum samples were isolated through centrifugation. Serum exosomes were isolated using an exosome isolation kit (4478360, Invitrogen, Massachusetts, USA). As for the tissues, potential RNA degradation was prevented by immediately immersing the specimens in an RNA preservation solution (R0118, Beyotime, Shanghai, China). Serum blood samples were harvested, aliquoted, and stored at −80°C. Patient data for the tissue samples are shown in Table 1.
| No | Age | Sex | Diagnosis | Metastasis | Tumor size (diameter, cm) | Stage |
|---|---|---|---|---|---|---|
| 1 | 61 | M | Squamous cell carcinoma | Absent | ≤3 | IB |
| 2 | 59 | M | Squamous cell carcinoma | Absent | >3 | IIA |
| 3 | 67 | F | Bronchioloalveolar carcinoma | Absent | ≤3 | IA |
| 4 | 47 | F | Adenosquamous carcinoma | Present | >3 | IIIB |
| 5 | 64 | M | Adenocarcinoma | Absent | >3 | IIB |
| 6 | 64 | M | Squamous cell carcinoma | Absent | >3 | IIA |
| 7 | 51 | F | Small-cell carcinoma | Present | >3 | IVB |
| 8 | 48 | M | Squamous cell carcinoma | Absent | ≤3 | IB |
| 9 | 62 | M | Adenocarcinoma | Absent | >3 | IIA |
| 10 | 63 | F | Mucoepidermoid carcinoma | Present | >3 | IIIA |
| 11 | 59 | M | Mucoepidermoid carcinoma | Absent | ≤3 | IB |
| 12 | 71 | M | Squamous cell carcinoma | Present | >3 | IIIA |
| 13 | 52 | M | Squamous cell carcinoma | Absent | >3 | IIA |
| 14 | 66 | F | Adenocarcinoma | Absent | ≤3 | IA |
| 15 | 65 | F | Small-cell carcinoma | Absent | ≤3 | IB |
| 16 | 41 | M | Adenocarcinoma | Absent | ≤3 | IB |
| 17 | 53 | M | Mucoepidermoid carcinoma | Absent | >3 | IIA |
| 18 | 43 | M | Mucoepidermoid carcinoma | Absent | ≤3 | IB |
| 19 | 48 | M | Bronchioloalveolar carcinoma | Present | >3 | IIB |
| 20 | 59 | M | Bronchioloalveolar carcinoma | Present | >3 | IIB |
Cell culture and co-culture
Human bronchial epithelial cell line (Bronchial Epithelial Cell Line 2B [BEAS2B], Delf-10130) and 95D cell line (Delf-10140) were newly purchased from the Hefei Wanwu Biotechnology Co., LTD; non-small-cell lung cancer cell lines A549 (1101HUM-PUMC000002), (Human lung cancer cell line 827, 1101HUM-PUMC000478), (National Cancer Institute-H1299, 1101HUM-PUMC000469), and (National Cancer Institute-H1650, 1101HUM-PUMC000251); lung squamous carcinoma cell line (Squamous Cell Carcinoma-MES 1, 1101HUM-PUMC000262); and small-cell lung cancer cell line (National Cancer Institute-H1688, 3101HUMTCHu154) were purchased from the National Infrastructure of Cell Line Resource of China (Beijing, China). A certificate of analysis confirming cell line identity through short tandem repeat profiling and confirming lack of mycoplasma contamination was obtained. The cells were cultured under 37°C in an environment with saturated humidity and 5% Carbon dioxide (CO2) in a Roswell Park Memorial Institute (RPMI) 1640 medium (C0893, Beyotime, Shanghai, China) enriched with 10% Fetal Bovine Serum (FBS; C0252, Beyotime, Shanghai, China) and 100 U/mL penicillin–streptomycin (C0222, Beyotime, Shanghai, China). Appropriate nutrition and pH balance were ensured by replacing the medium every 2–3 days.
Interactions between lung cancer and endothelial cells were evaluated with a Transwell chamber co-culture system (3422, Corning, New York, USA). Lung cancer cells were seeded in the bottom chamber, and TAECs were positioned in the top chamber in a ratio of 5:1. Afterward, the chambers were incubated under standard culture conditions: temperature of 37°C, 100% humidity, and atmosphere containing 5% CO2.
TAEC isolation
TAECs were isolated from resected lung carcinoma specimens obtained immediately after surgical removal.[20] The specimens were meticulously minced and enzymatically dissociated in an RPMI 1640 medium enriched with 0.1% collagenase IV (C5138, Sigma–Aldrich, Missouri, USA) at 37°C for 1 h. The resultant cellular suspension was sequentially filtered through a series of graded meshes, and stromal and aggregated materials were discarded. TAECs were enriched using anti-CD34 monoclonal antibodies (ab81289, Abcam, Massachusetts, USA) bound to magnetic beads, and isolation was carried out with a magnetic cell sorting (MACS) system (Miltenyi Biotech, Bergisch Gladbach, Germany). Diphtheria toxin (500 ng/mL, Calbiochem, San Diego, CA, USA) was added to the TAEC cultures to eliminate remaining human tumor cells. After 24 h, dead cells were aspirated, and non-endothelial tumor cells were completely removed. The TAECs were further purified using a second MACS step, and Fluorescein Isothiocyanate-labeled BS1-B4 Lectin (FITC-BS1-B4 lectin) was used to eliminate contaminating stromal cells and enhance the purity of the endothelial cells. The purity of the isolated TAECs was confirmed through flow cytometric (FCM) analysis. Markers, such as FITC-BS1-B4, were used to ensure that the cell population was primarily composed of endothelial cells. After isolation, the TAECs were tested and found negative for mycoplasma contamination. The TAECs were grown in endothelial growth medium-2 (CC-3162, Lonza Group Ltd., Basel, Switzerland), which was enhanced with 10% FBS and 100 U/mL penicillin–streptomycin. The cultured TAECs were then passaged at subconfluency, and experiments were conducted using cells from passages 1–6 to ensure cellular fidelity.
Exosome extraction and characterization detection
After tumor cells were transfected with miR-5703 mimics, inhibitors, and negative control (NC) (GenePharma, Suzhou, China), the culture supernatant was collected and retained after 10 min of centrifugation at ×350 g. Then, the supernatant was centrifuged again (×2000 g for 30 min). The supernatant was mixed with an exosome extraction working solution (C10130-1, Ribo Bio, Guangzhou, China) and left overnight at 4°C. The next day, the samples were centrifuged, and the lower sediment was retained. The exosomes were reconstituted in an optimal volume of Phosphate-Buffered Saline (PBS) and transferred to a cuvette. Exosome particle size analysis was carried out using a nanoparticle size analyzer (N30E, Xiamen Fuliu Biological Technology Co., Ltd, Xiamen, China). Exosome markers (CD81, CD63, and Alix) and negative markers (calnexin and α-tubulin) were detected by Western blotting. In addition, CD81 (ab79559, Abcam, Massachusetts, USA) was confirmed by FCM.[21] The method for isolating exosomes from patient sera was the same as that used for isolating exosomes from cell supernatants.
Exosomes were isolated and then fixed using 100 μL of 2.5% glutaraldehyde (G1102, Servicebio, Wuhan, China) and kept at 4°C. An exosome suspension (5 μL) was gently placed on a copper grid, where the exosomes were allowed to adhere for 20 min at room temperature. Subsequently, the grids were rinsed with PBS and then fixed with 1% glutaraldehyde for 5 min. The samples were rinsed 10 times with double- distilled water and negatively stained with 4% uranyl acetate (Hubei Xingyinhe Chemical Co., Ltd. Wuhan, China) for 5 min. Any excess staining solution was carefully dabbed off with filter paper, and the samples were allowed to dry at ambient temperature. The exosomes were visualized with a transmission electron microscope (H-7000; Hitachi, Ltd., Tokyo, Japan). The reagents, including the culture medium, FBS, and liquid used in this experiment, were exosome free.
Transwell assay
TAECs were seeded into six-well plates at a density of 2 × 105 cells per well and cultured until they reached roughly 70% confluency. The isolated exosomes were then added to the TAECs at a concentration of 50 μg/mL, and the culture was maintained for 24–48 h at 37°C in a 5% CO2 atmosphere. Subsequently, a cell suspension consisting of either endothelial cells or lung cancer cells was prepared at a concentration of 1 × 106 cells/mL. This cell suspension was then added to the Transwell migration chamber (3428, Corning, New York, USA), typically dispensing 500 μL per well. The lower chamber was either seeded with an alternate cell type (TAECs or lung cancer cells) or left cell free. The Transwell chamber was incubated at 37°C with 100% humidity. After 48 h of incubation, the upper chamber was collected. Cells that did not migrate from the bottom of the inner chamber were scraped off, retaining only the cells that had migrated through the membrane. The cells were visualized through underwent 10 min 2% crystal violet staining, which were subsequently counted and imaged under a microscope. The migration behavior of the cells was then analyzed. To assess the invasive capability of lung cancer cells, we added a layer of Matrigel matrix (C0372, Beyotime, Shanghai, China) to the bottom of the Transwell chamber. On the basis of the number of cells that penetrated this matrix, the invasive potential of the cells was evaluated.
Cell proliferation assay
Cell viability was determined using a cell counting kit 8 (CCK8) kit (CCK-8, Dojindo, Kumamoto, Japan) and 5-ethynyl-2'-deoxyuridine (EdU) staining. After the cells were transferred with miRNA mimics, inhibitors, or plasmids, the cells were prepared for cell viability detection with the CCK8 kit or EdU staining.
CCK8
Cell viability was assessed through a CCK-8 assay. Log-phase cells were harvested and resuspended to achieve a density of 10,000 cells/well in a 96-well plate. Each well was filled with 100 μL of the cell suspension. The plate was then incubated at 37°C in a 5% CO2 atmosphere for 24 h. After incubation, 10 μL of a CCK-8 solution was introduced to each well, and the plate was further incubated for 1 h. Finally, the absorbance of each well was measured at 450 nm with a microplate reader (Thermo Fisher Scientific, mμltiscan MK3).
EdU staining
Cell proliferation was assessed using an EdU staining kit (C0075L, Beyotime, Shanghai, China). The cells were treated with 50 μM EdU for 2 h at 37°C, fixed with 4% paraformaldehyde (G1101, Servicebio, Wuhan, China), and permeabilized with 0.3% Triton X-100 (GC204003, Servicebio, Wuhan, China). Subsequently, the cells were exposed to a click reaction mixture for 30 min in accordance with the manufacturer’s guidelines. Finally, the cells were stained with 4',6-diamidino-2'-phenylindole (DAPI, C1002, Beyotime, Shanghai, China), and the proportion of EdU- positive cells, which is indicative of proliferating cells, was assessed through fluorescence microscopy (Eclipse 80i, Nikon, Tokyo, Japan).
Cell apoptosis detection
Hoechst: Initially, cover slips were sterilized in 70% ethanol for at least 5 min and then air-dried or rinsed 3 times with PBS. After washing with the cell culture medium, cover slips were placed in six-well plates, and the cells were seeded and grown to 50–80% confluency. To measure cell apoptosis, we replaced the medium with 0.5 mL of fixative. After fixation, the cover slips were washed twice with PBS for 3 min each. The cells were then stained with 0.5 mL of Hoechst 33258 dye (C1011, Beyotime, Shanghai, China) for 5 min, agitated, and washed twice. Subsequently, the cells were stained with 0.5 mL of DAPI staining solution (C1002, Beyotime, Shanghai, China) for 5 min and washed twice. An antifade mounting medium was applied for microscopy. The stained cell nuclei appeared blue under a fluorescence microscope at excitation and emission wavelengths of approximately 350 and 460 nm, respectively.
Flow Cytometry (FCM)
Cells were digested, centrifuged at ×500 g for 5 min at 4°C, and washed twice with PBS. Afterward, the cells were resuspended in ×1 binding buffer to a final volume of 5 × 106/mL, and 100 μL of the suspension was treated with 5 μL of annexin V–fluorescein isothiocyanate (C1062S, Beyotime, Shanghai, China) and 5 μL of propidium iodide (PI, C1062S, Beyotime, Shanghai, China). The suspension was then incubated in the dark at room temperature for 8–10 min. After 400 μL of ×1 binding buffer was added, the mixture was analyzed through FCM within an hour. The apoptosis rate was determined by the sum percentages of annexin V+/PI+ and annexin V+/PI- cells.
Immunofluorescence assay
After co-cultivation with lung cancer cells, the expression of α-smooth muscle actin (α-SMA) and ING4 in endothelial cells was evaluated through immunofluorescence. Fixed with 4% paraformaldehyde for 15 min. Then, the cells were infiltrated with 0.1% Triton X-100 for 5 min and blocked with 1% BSA at room temperature for 1 h. Primary antibodies targeting ING4 (1:200, ab113425, Abcam, Massachusetts, USA) and (α-SMA, 1:100, ab7817, Abcam, Massachusetts, USA) were applied to the cells, which were incubated overnight. Then, the cells were incubated with suitable secondary antibodies (1:1000, ab150077, Abcam, Massachusetts, USA). Finally, the cells were stained with DAPI (C1002, Beyotime, Shanghai, China) and examined using a confocal laser scanning microscope (OLS5000, Olympus, Tokyo, Japan). The expression levels of ING4 and α-SMA in the endothelial cells were examined using images.
Tube formation assay
Endothelial cells, on reaching 80% confluency, were washed with PBS once or twice and then treated with trypsin (P4201, Beyotime, Shanghai, China). They were then incubated at 37°C for 2–3 min. To neutralize trypsin, a fresh complete medium was introduced, and the cells were resuspended to yield a single-cell suspension. The cell density was adjusted to 1 × 106 cells/mL. Subsequently, 100 μL of the cell suspension with 1 × 105 cells/mL was plated into a six-well plate and cultured overnight at 37°C in a 5% CO2 atmosphere. The next day, after specific treatments, the cells were incubated for 48 h. Concurrently, Matrigel was thawed overnight at 4°C. All materials, including pipettes and tips, were prechilled. With a cold pipette tip, 100 μL of Matrigel was added to a prechilled 48-well plate, and even distribution in the wells was ensured. After the application of Matrigel to the plate, the plate was incubated for 40 min at 37°C for the solidification of the Matrigel matrix. Subsequently, the cells were counted, and their density was adjusted to 3 × 105 cells/mL. The Matrigel-coated 48-well plate was washed with PBS 3 times. The volume of 200 μL of the cell suspension was 6 × 104 cells. The suspension was dispensed into each well until the density was 8 × 104 cells per well. The plate was then maintained at 37°C in an atmosphere containing 5% CO2. After 5 h, tube formation was inspected using an inverted microscope, and images were obtained for subsequent analysis.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from the collected clinical tissues and cell lines with TRIzol reagent (15596026, ThermoFisher, Illinois, USA). A reverse transcription kit (G3335-50, Servicebio, Wuhan, China) was used in synthesizing complementary DNA (cDNA). For qRT-PCR detection, a reaction mixture consisting of cDNA, SYBR Green fluorescent dye and specific primers was prepared using SYBR Green I Master Mix on a 7900 sequence detector (Applied Biosystems, Foster City, CA, USA). The conditions for qRT- PCR were as follows: Initial denaturation step at 95°C for 10 min for 40 cycles. Each cycle consisted of a denaturation phase at 95°C for 15 s and an annealing/extension phase at 60°C for 1 min. The data were analyzed using the 2−ΔΔCT method.[22] For miRNAs, U6 served as the internal reference. As for mRNAs, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as the internal reference. The primer sequences in RT-PCR are provided in Table S1.
Western blot
Protein lysates from tissues or cells were prepared using a radio-immunoprecipitation assay buffer (G2002, Servicebio, Wuhan, China) containing protease and phosphatase inhibitors (G2006, Servicebio, Wuhan, China). Protein concentrations were measured with a bicinchoninic acid assay. Equal amounts of the proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (G6015, Servicebio, Wuhan, China). These were incubated overnight at 4°C with primary antibodies against E-cadherin (1:1000; ab227639; Abcam), N-cadherin (1:1000; ab76011; Abcam), vimentin (1:1000; EPR3776; Abcam), Matrix Metalloproteinase-2 (MMP2, 1:1000; ab92536; Abcam), Matrix Metalloproteinase-9 (MMP9, 1:1000; ab76003; Abcam), and GAPDH (1:1000; ab8245; Abcam). After incubation with primary antibodies, the membranes were washed and treated with Horseradish Peroxidase -linked secondary antibodies (1:5000, ab205718, Abcam, Massachusetts, USA), and bands were detected using an enhanced chemiluminescence system (G2020, Servicebio, Wuhan, China). Band intensities were quantified, and expression was normalized to that of GAPDH.
In vivo model
Breeders’ Association of Laboratory Animals, California nude mice (6–8 weeks old, approximately 20 g, n = 20) were purchased from Animal Center of Tianjin Medical University. They were subcutaneously injected with 5 × 106 H1299 cells each. The mice were divided into four groups (n = 5) and received intratumoral injections every 3 days: 100 μL PBS (the blank group), H1299-derived exosomes (the control group), exosomes with miR-5703 inhibitor NC (the NC group), and exosomes with miR-5703 inhibitor (the inhibitors group). Tumor volumes were measured weekly with the formula V = width2 × length/2 (mm3). On day 28, mice were euthanized after 1% pentobarbital sodium anesthesia (50 mg/kg). Tumors were collected, weighed, and prepared for histological analysis. For hematoxylin and eosin (H&E) staining, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), and immunohistochemistry, the tissues were fixed in 4% paraformaldehyde. For quantitative PCR (qPCR) and Western blot, the samples were stored at −80°C. Ethics approval was obtained from the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (approval No. 2023078). All animal experiments were conducted in strict accordance with the guidelines for the care and use of laboratory animals established the Second People’s Hospital of Yibin. All procedures were performed under appropriate anesthesia, and all animals received humane care throughout the duration of the study.
H&E staining
The tissues were fixed in paraformaldehyde, embedded in paraffin, and sectioned continuously (4 μm). After dehydration and dewaxing, the sections were sequentially stained with H&E (G1076, Servicebio, Wuhan, China), mounted with neutral resin, and examined under a bright- field microscope (Eclipse 80i, Nikon, Tokyo, Japan) for histopathological analysis.
Immunohistochemistry
After dehydration and dewaxing, the tissue sections were subjected to antigen retrieval and peroxidase blocking. After blocking with serum, the sections were incubated overnight with the following primary antibodies: E-cadherin (1: 100, ab227639; Abcam), N-cadherin (1: 50, ab76011, Abcam), Ki67 (1:500, ab15580; Abcam), and CD34 (1:200, ab81289; Abcam). Then, the sections were incubated with secondary antibodies for 1 h (1:5000, ab205718, Abcam). After developing with 3,3’-Diaminobenzidine (G1212, Servicebio, Wuhan, China), the sections were counterstained with hematoxylin, mounted with neutral resin, and examined under a bright-field microscope (Eclipse 80i, Nikon, Tokyo, Japan) for the assessment of protein expression levels and localization.
TUNEL assay
After dehydration and dewaxing, the tissue sections were treated with proteinase K (MK1012-100, Boster Biotechnology, Wuhan, China). After thorough washing with PBS, a TUNEL assay reagent consisting of Terminal Deoxynucleotidyl Transferase enzyme, fluorescence labeling solution, and TUNEL detection solution (MK1012-100, Boster Biotechnology, Wuhan, China) was applied. The sections were then incubated at 37°C for 60 min, extensively washed with PBS, and finally mounted using an antifade mounting medium to prevent fluorescence quenching. The sections were then observed with a fluorescence microscope (Eclipse 80i, Nikon, Tokyo, Japan). The excitation and emission wavelengths of Cy3 were 550 and 570 nm, respectively, resulting in red fluorescence.
Luciferase assay
For ING4 and has-miR-5703 luciferase assay, the cells were co-transfected with 50 nM miR-5703 mimics and 200 ng of a pmirGLO-ING4 vector or corresponding mutant type. A dual luciferase assay kit (Promega, WI, USA) was used for dual luciferase assay. At 48 h post-transfection, cells were collected, and Renilla luciferase activity was assessed. Results were assessed as the ratios of Firefly luciferase activity to Renilla luciferase activity.
Data collection and bioinformatics analysis
To explore the potential mechanisms of circulating exosomal biomarkers in the plasma of patients with lung cancer, we selected the GEO Series 198959 (GSE198959) dataset to analyze the differentially expressed miRNAs in the Exos of patients with recurrent lung cancer. At an adjusted P < 0.05 and |log2 (Fold Change)| of >1, highly expressed miRNAs were screened for subsequent validation. Kaplan–Meier curves were generated using Kaplan–Meier Plotter (KMplot, www.kmplot.com).[23] The gene expression data and relapse- free and Overall Survival information in the KMplot program were downloaded from Gene Expression Omnibus (GEO), European Genome-phenome Archive, and The Cancer Genome Atlas.
Data analysis
Data were presented as mean ± Standard deviation. The Statistical Package for the Social Sciences 19.0 (IBM, Armonk, NY, USA) was used for statistical analysis. Analysis of variance and Turkey’s post hoc tests were used for multiple- group comparisons, and t-test was utilized for two-group comparisons. P < 0.05 indicated statistical significance. Image editing was conducted using GraphPad Prism 9.0 (GraphPad Software Inc., La Jolla, CA, USA).
RESULTS
Expression characteristics of miR-5703 in lung cancer
In a bid to identify potential factors in lung cancer, a study was conducted based on the GEO dataset database, using the GSE198959 dataset. Five miRNAs, namely, miR-4286, miR-6869, miR-630, miR-5703, and miR- 7641, were identified. By performing qPCR detection on clinical lung cancer tissues (T) and peri-cancerous tissues, it was observed that the expression levels of the miRNAs significantly varied in the lung cancer tissues (P < 0.0001), [Figure 1a].
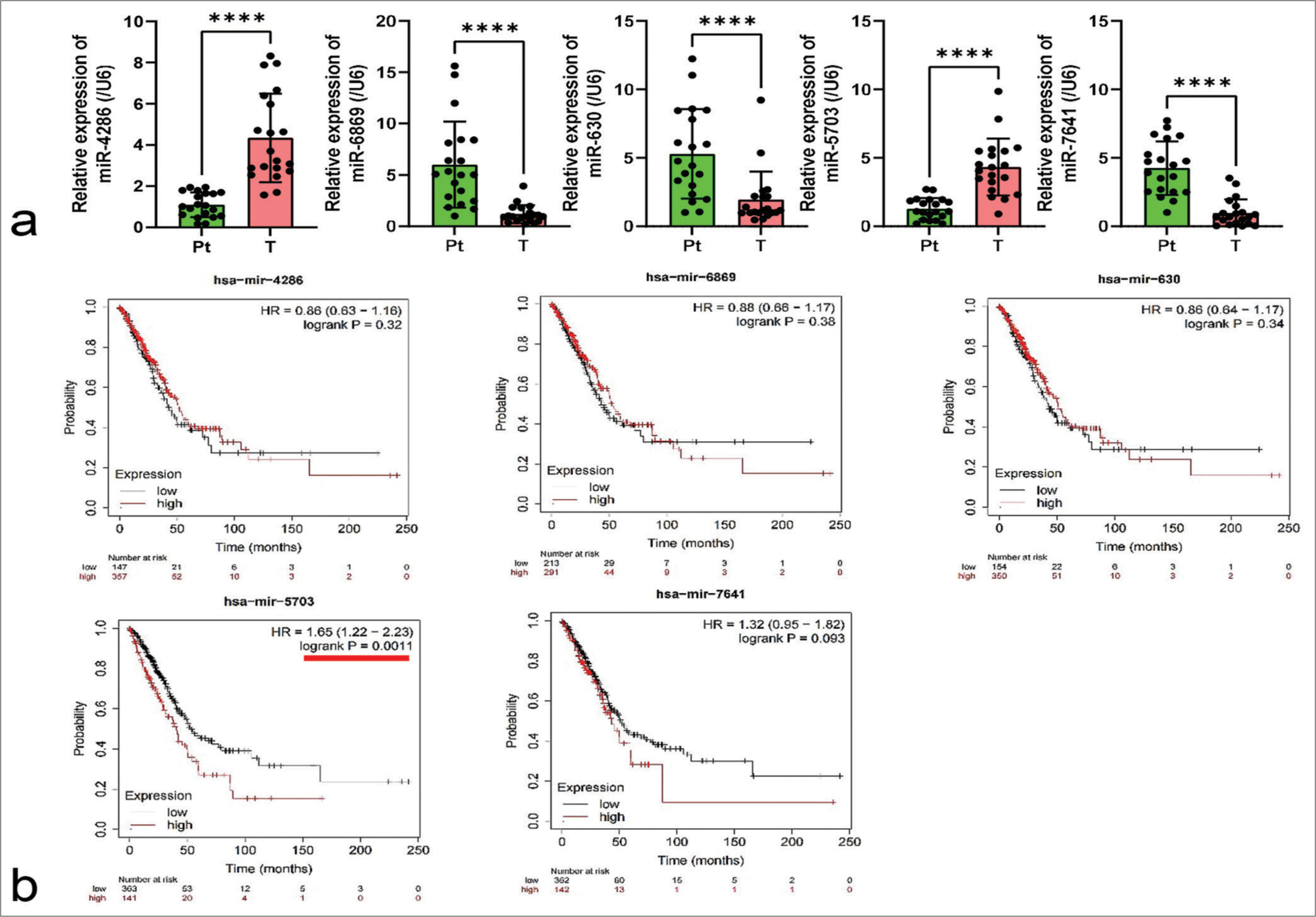
- Expression profiling and clinical relevance of microRNA-5703 (miR-5703) in lung cancer. (a) Quantitative polymerase chain reaction detection of the relative expression levels of miR-4286, miR-6869, miR-630, miR-5703, and miR-7641 in lung cancer tissues compared with peri-cancerous tissues. Significant difference in the expression of each microRNA was found between the groups. (b) Kaplan– Meier survival analysis revealed the significant correlations of miR-4286, miR-6869, miR-630, miR-5703, and miR-7641 expression levels to patient prognosis. n = 20, ✶✶✶✶P < 0.0001 versus peri-cancerous tissues. Pt: peri-cancerous tissues; T: lung cancer tissues
Further, employing the Kaplan–Meier Plotter platform (http://kmplot.com/analysis/index.php?p=background), the correlation of miR-4286, miR-6869, miR-630, miR-5703, and miR-7641 with lung cancer patients was respectively analyzed. Results revealed a significant correlation between miR-5703 and patient prognosis (P = 0.0011), [Figure 1b].
Expression characteristics of miR-5703 in exosomes of patients with lung cancer
Exosomes were isolated from the serum samples of healthy individuals and patients with lung cancer for characterization and detection. Electron microscopy observation showed that the exosomes were approximately 100 nm in size [Figure 2a]. Western blotting revealed the presence of CD81, CD63, and Alix and the absence of calnexin and α-tubulin within the exosomes [Figure 2b]. FCM tests showed a clear CD81-positive marker in the exosomes [Figure 2c].
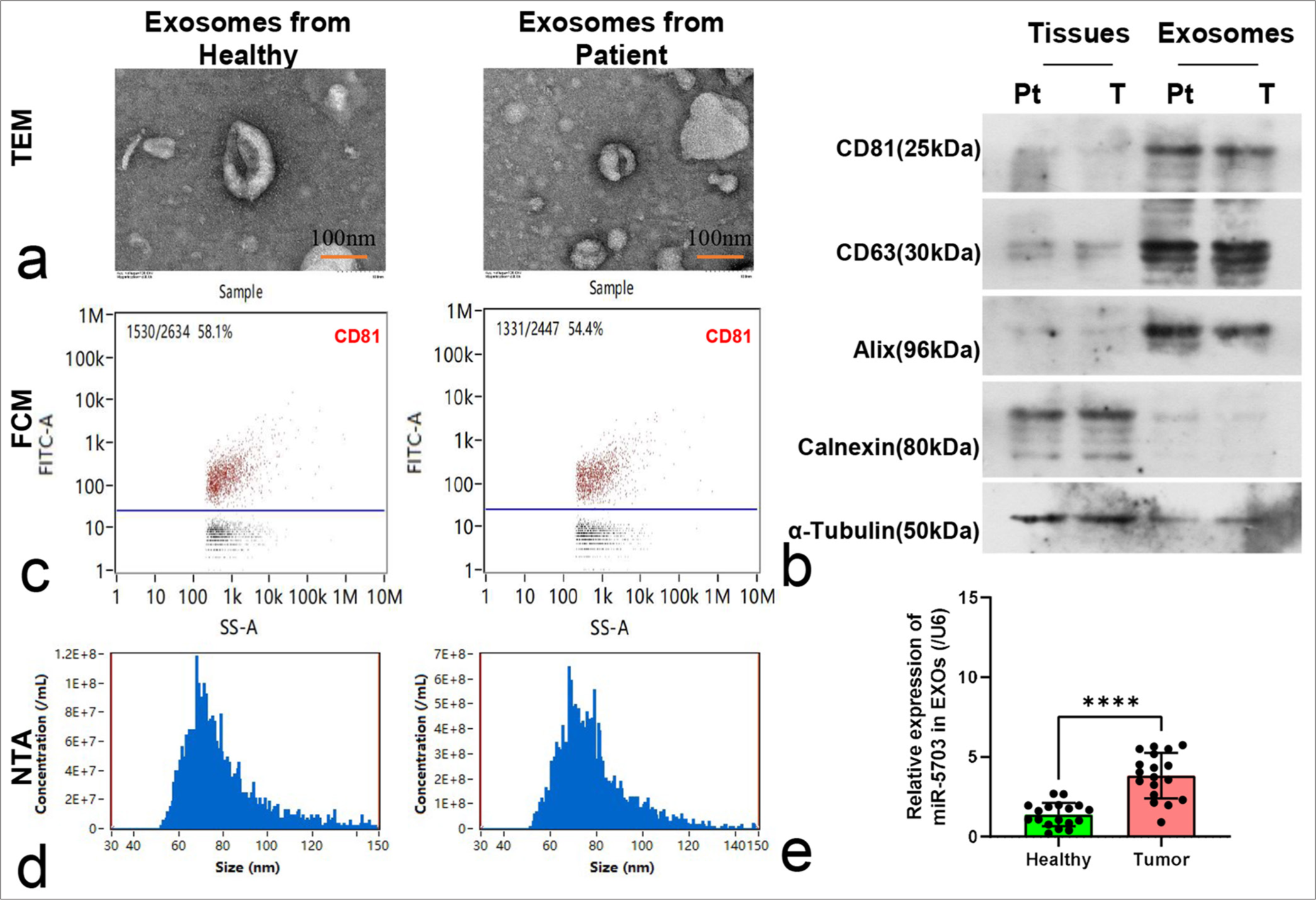
- Characterization and microRNA-5703 (miR-5703) quantification of exosomes derived from serum of lung cancer patients and healthy individuals. (a) Electron microscopy image showing exosomes of approximately 100 nm in size, n = 3. (b) Western blotting results of specific exosome markers CD81, CD63, and Alix and negative markers calnexin and α-tubulin, n = 3. Pt: healthy individuals; T: lung cancer patients. (c) Flow cytometry test showing the CD81-positive marker within the exosomes, n = 3. (d) Particle size analysis of the extracted exosomes showing diameters ranging from 50 nm to 120 nm, n = 3. (e) Quantitative polymerase chain reaction results showing significantly higher levels of miR-5703 in the serum exosomes of patients with lung cancer compared with the healthy individuals, n = 20. ✶✶✶✶P < 0.0001 versus health control. SS-A: Side scatter-area
Subsequent analysis of particle size revealed that the diameters of the isolated exosomes varied from 50 nm to 120 nm, and no significant difference in diameter was observed between the exosomes obtained from the healthy individuals and those from patients with lung cancer [Figure 2d]. However, qPCR results showed that the concentration of miR-5703 in serum exosomes from the patients with lung cancer was significantly elevated compared with that in serum exosomes from the healthy individuals (P < 0.0001), [Figure 2e].
Biological role of miR-5703 in lung cancer cells
To explore the biological role of miR-5703 in lung cancer, the experiment incorporated various lung cancer cell lines and a non-tumorigenic cell line (BEAS2B). The highest expression levels of miR-5703 were observed in the 95-D and H1299 cell lines [Figure 3a]. Thus, these cell lines were selected for further experiments.
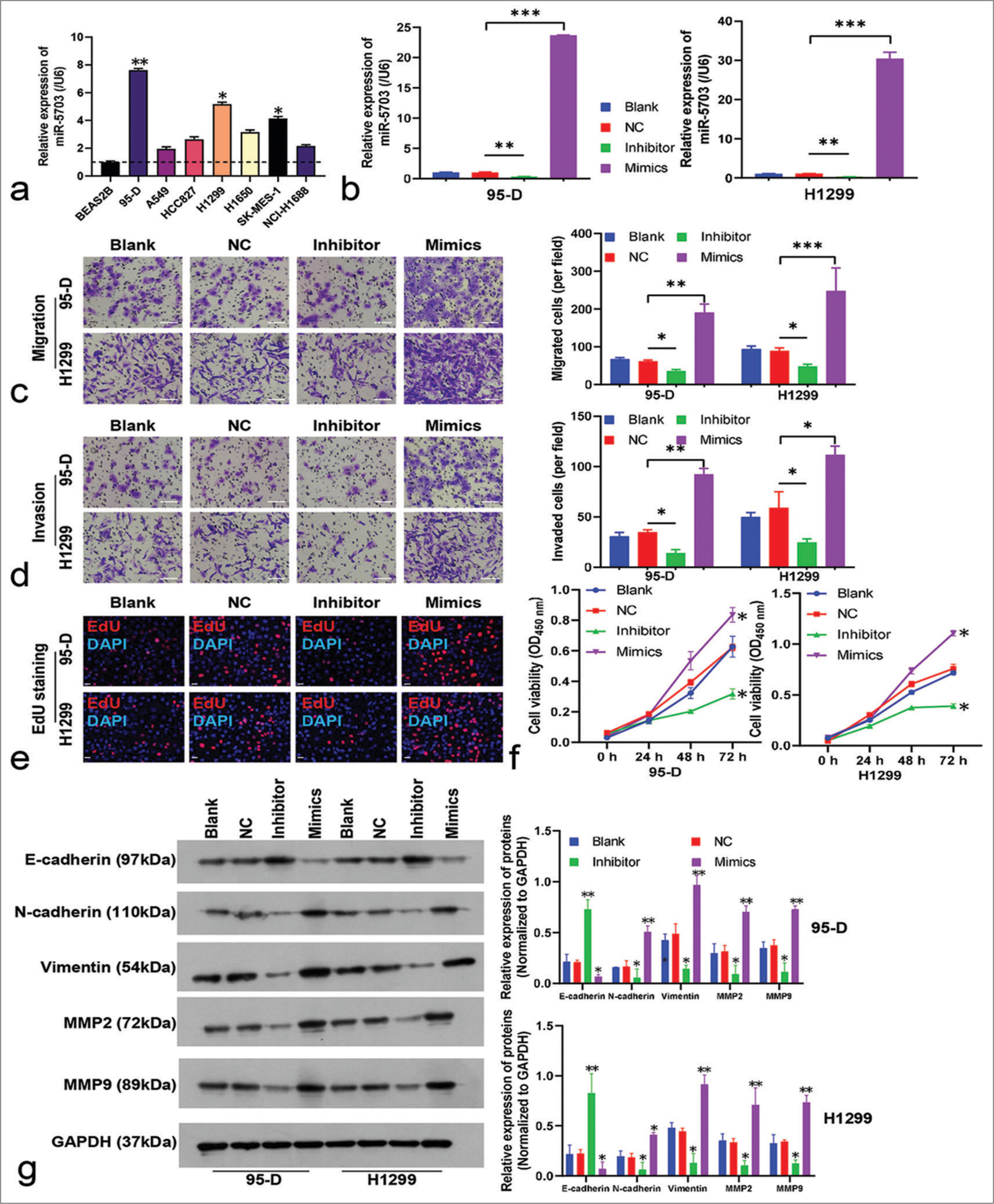
- Functional analysis of microRNA-5703 (miR-5703) in 95-D and H1299 lung cancer cell lines. (a) Relative miR-5703 expression levels in different lung cancer cell lines and non-tumor cell line Bronchial Epithelial Cell Line 2B as detected by quantitative polymerase chain reaction (qPCR).SK-MES: Squamous Cell Carcinoma- MES; NCI-H1688: National Cancer Institute- H1688. (b) miR-5703 expression in the 95-D and H1299 cell lines after treatment with miR-5703 inhibitors or mimics, as demonstrated by quantitative polymerase chain reaction. (c and d) Cell migration (c) and invasion (d) assays showing the effects of miR-5703 expression modulation in the 95-D and H1299 cells. Scale: 50 μm. (e and f)5-ethynyl-2'-deoxyuridine staining (e, scale: 20 μm) and CCK8 kit method tests. DAPI: 4’,6-Diamidino-2-phenylindole, a commonly used fluorescent dye for staining the nucleus. (f) demonstrating changes in cell proliferation and vitality respective to different treatments. (g) Western blot results showing altered the expression of E-cadherin, N-cadherin, vimentin, MMP2, and MMP9 in the cells treated with the miR-5703 inhibitors or mimics. MMP2: Matrix Metalloproteinase-2; MMP9: Matrix Metalloproteinase-9; NC: Negative Control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase. n = 3, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001 versus negative control.
In the 95-D and H1299 cells, the expression of miR-5703 was manipulated. As shown in Figure 3b, the inhibitors led to a significant decrease in miR-5703 expression (P < 0.01), whereas the mimics caused a dramatic increase in miR- 5703 expression (P < 0.001), [Figure 3b]. Subsequently, the malignant behavior of the cells was assessed. As shown in Figure 3c and d, the inhibitors led to a significant decrease in 95-D and H1299 cell migration and invasion (P < 0.05), whereas the mimics led to an increase in migration and invasion (P < 0.05), [Figure 3c and d]. The results of EdU staining and CCK8 kit on the 95-D and H1299 cells showed that cell proliferation was significantly inhibited and promoted, respectively (P < 0.05), [Figures 3e and f, S1a].
Western blot results showed that the inhibitors enhanced the expression of E-cadherin and suppressed the expression of N-cadherin, vimentin, MMP2, and MMP9. Meanwhile, cells treated with the mimics showed reduced E-cadherin and increased N-cadherin, vimentin, MMP2, and MMP9 expression levels [Figure 3g].
Consistent with the FCM results, Hoechst staining results showed that lung cancer cells treated with the inhibitors had increased number of apoptosis cells (P < 0.05), [Figure 4a and b]. Conversely, after the cells were treated with the mimics, apoptosis rate decreased significantly (P < 0.05), [Figure 4a and b]. These results suggested that miR-5703 promoted the malignant biological behavior of the lung cancer cells. Notably, after treatment with the miR-5703 inhibitors and mimics, the exosomes showed a significant decrease (P < 0.01) or increase (P < 0.001) in miR-5703 abundance, respectively [Figure 5].
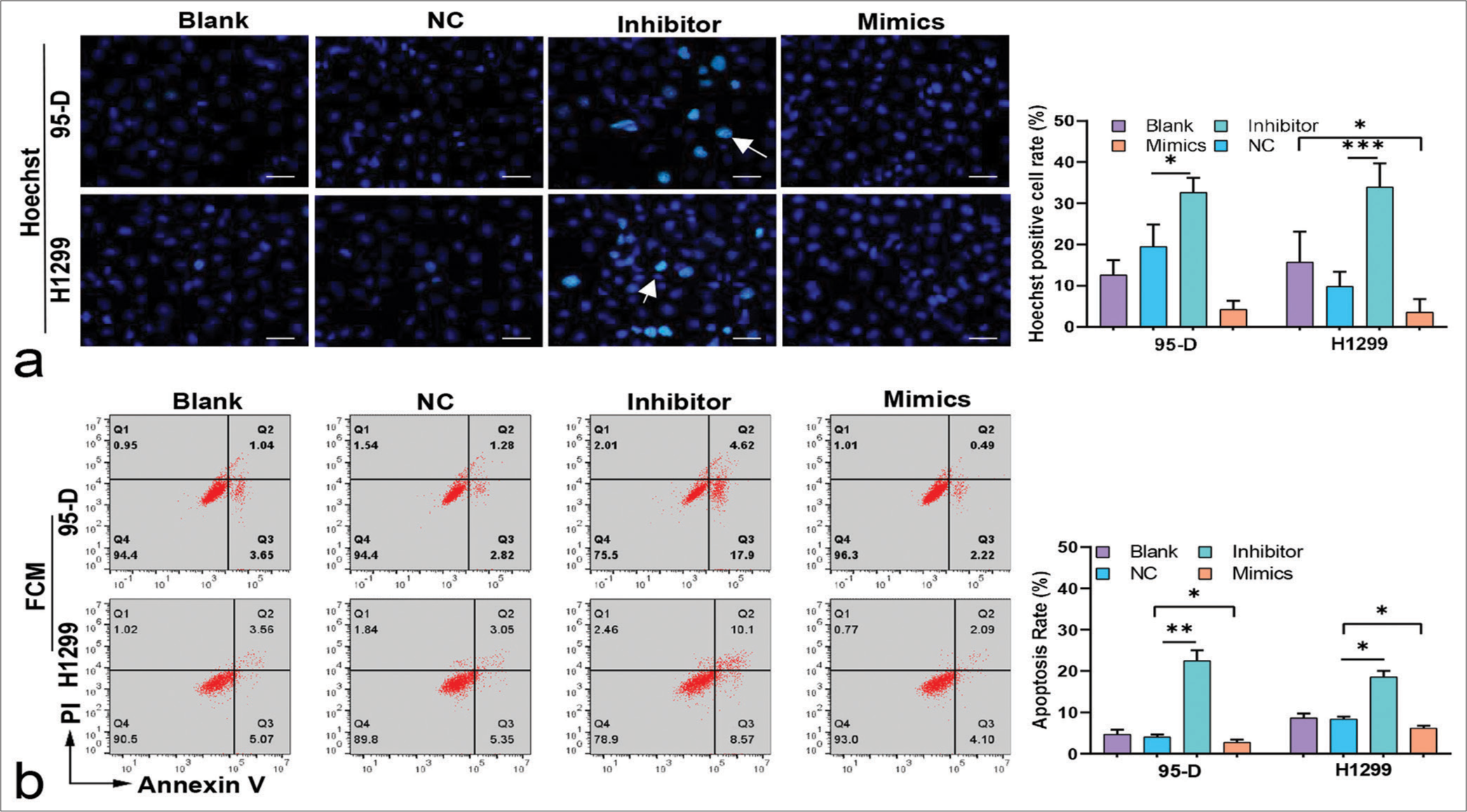
- Impact of microRNA-5703 (miR-5703) modulation on apoptosis in lung cancer cells. (a) Hoechst staining images showing increased apoptosis in lung cancer cells treated with the miR-5703 inhibitors and decreased apoptosis after treatment with mimics. White arrows indicate apoptotic cells. Scale: 20 μm. (b) Flow cytometry results reflecting those of Hoechst staining showing change in apoptosis levels after the treatments. n = 3, ✶P < 0.05, ✶✶P < 0.01. NC: Negative Control.
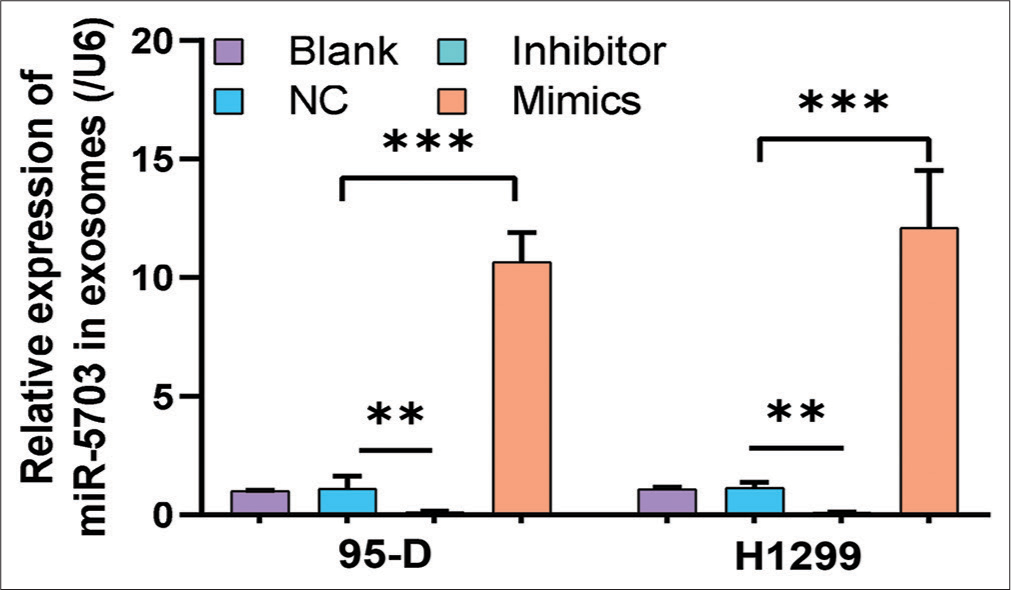
- Modulation of microRNA-5703 (miR-5703) levels in exosomes derived from lung cancer cells after treatment with inhibitors and mimics. Quantitative polymerase chain reaction results showing a significant decrease in miR-5703 expression in exosomes derived from lung cancer cells treated with the inhibitors and significant increase after treatment with mimics. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. NC: Negative Control.
Effect of lung cancer cell–derived exosomes on TAECs
TAECs isolated from lung cancer tissues were CD34 positive according to the immunofluorescence [Figure 6a] and FCM results, and the positive ratio exceeded 95% [Figure 6b].
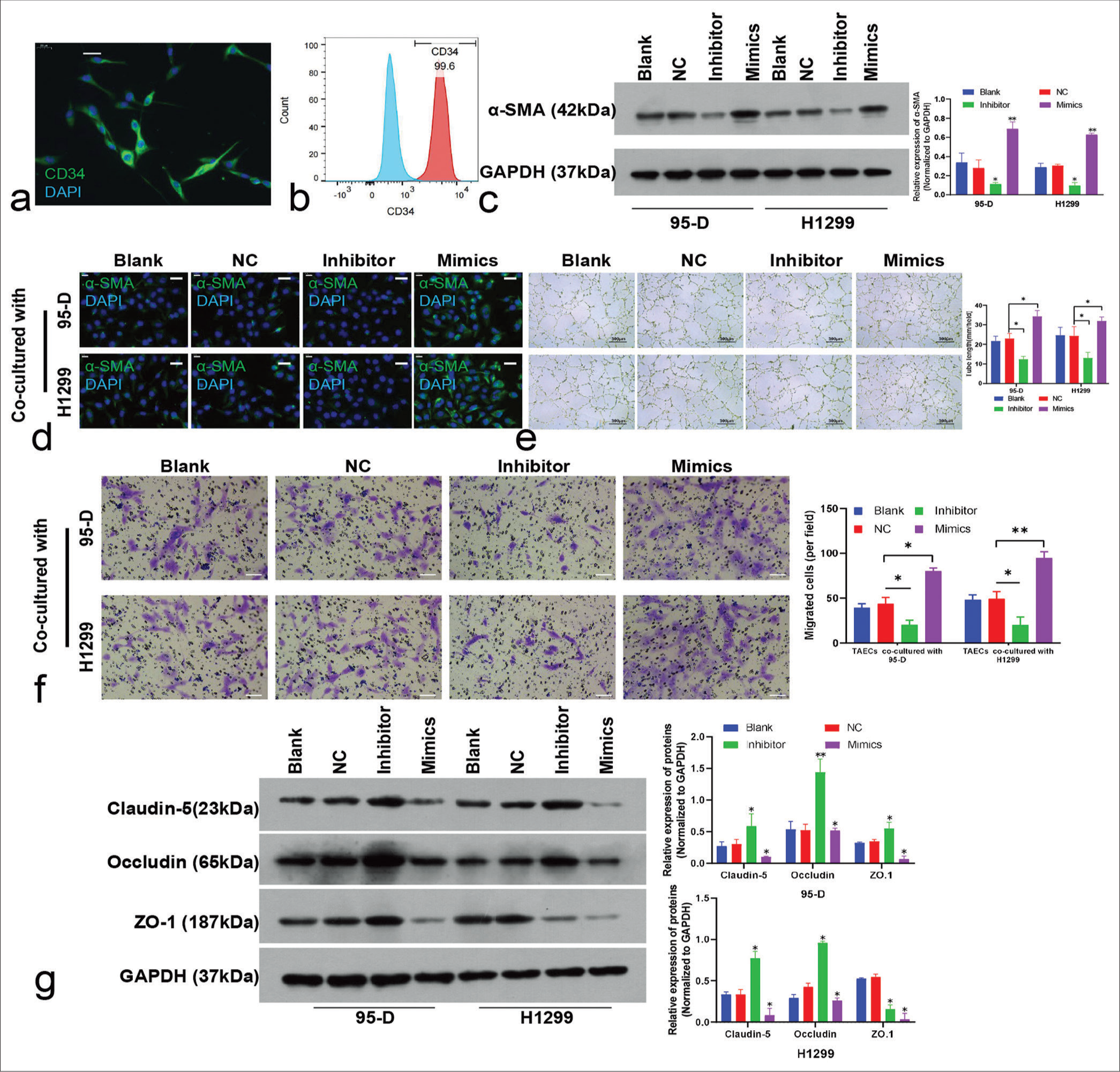
- Influence of microRNA-5703 (miR-5703) modulation in lung cancer cells on tumor-associated endothelial cells (TAECs). (a) Immunofluorescence visualization (scale: 20 μm) and (b) flow cytometry quantification revealing >90% CD34-positive ratio in isolated TAECs from lung cancer tissues. (c-d) Western blot (c) and immunofluorescence (d, scale: 20 μm) displayed change in α-smooth muscle actin (α-SMA) expression in endothelial cells upon co-culture with lung cancer cells treated with miR-5703 inhibitors or mimics. GADPH: glyceraldehyde-3-phosphate dehydrogenase. (e-f) Variation in the angiogenesis (e) and migration (f, scale: 50 μm) of endothelial cells after co-cultivation. (g) Western blot analysis of Claudin-5, Occludin and ZO-1 protein expression in endothelial cells co-cultured with lung cancer cells treated with inhibitors or mimics. n = 3, ✶P < 0.05, ✶✶P < 0.01 versus negative control. NC: Negative Control.
The co-cultivation of TAECs and lung cancer cells were performed. Notable changes in α-SMA expression in TAECs were observed. Western blot and immunofluorescence results showed that α-SMA expression was attenuated in the TAECs co-cultured with inhibitor-treated lung cancer cells. By contrast, endothelial cells co-cultured with lung cancer cells treated with the mimics showed rapid increase in α-SMA expression (P < 0.01), [Figure 6c]; (P < 0.05), [Figures 6d and S1b]. The results showed that TAECs co-cultured with lung cancer cells treated with inhibitors showed inhibited tube formation and migration. Conversely, lung cancer cells treated with mimics promoted TAECs tube formation and migration abilities (P < 0.05), [Figure 6e and f]. The elevated expression levels of claudin-5, occludin, and ZO-1 were observed in the TAECs co-cultured with inhibitor-treated lung cancer cells, and expression in TAECs was suppressed after co-culturing with mimic-treated lung cancer cells. Surprisingly, in H1299, ZO-1 expression was reversely reduced under inhibitor treatment (P < 0.05), [Figure 6g].
Lung cancer cell-derived exosomal miR-5703 promoted TAEC EndMT and angiogenesis and inhibited cell adjunction
To elucidate the effect of exosomal miR-5703 on the biological functions of TAECs, the exosomes derived from lung cancer cells were isolated, and then co-cultured with TAECs. Concurrently, to assess the direct impact of exosome uptake on miR-5703 levels in TAECs, the TAECs were treated with Cytochalasin D, an inhibitor known to block exosome uptake, as a NC to mimic the absence of exosomes. As shown in Figure 7a, PKH67-labeled exosomes were taken up by the TAECs. Exosomes derived from lung cancer cells significantly up-regulated miR-5703 levels in the TAECs (P < 0.001), but treatment with cytochalasin D reversed this effect (P < 0.001). No significant difference in miR-5703 level was observed between these cells and the cells in the blank group, indicating the effective inhibition of exosome uptake [Figure 7b].
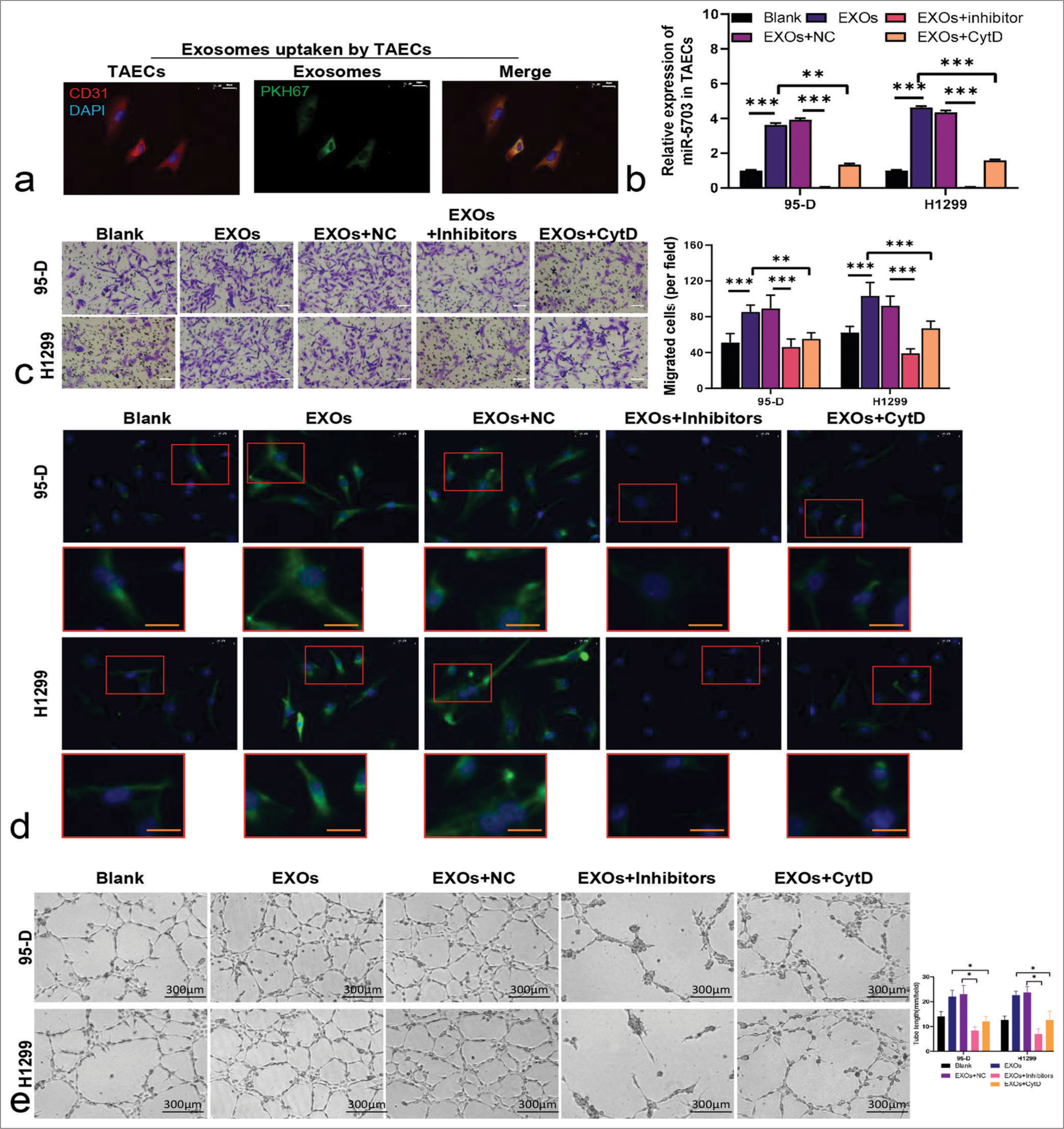
- Lung cancer cell-derived exosomes encapsulating microRNA-5703 (miR-5703) regulate EndMT, angiogenic ability, and cell adjunction in tumor-associated endothelial cells (TAECs). (a) Confocal microscopy images Ashowing PKH67-labeled exosome uptake by TAECs after co-culturing. Scale: 20 μm. (b) Quantitative polymerase chain reaction analysis revealing increased miR-5703 levels in TAECs co-cultured with exosomes or treated with cytochalasin D. (c) Migration assay demonstrating the enhanced migratory capacity of TAECs. Scale: 50 μm. (d) α-smooth muscle actin expression was measured through immunofluorescence. Scale: 20 μm. (e) The angiogenesis of TAECs was evaluated using tube formation assay. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. EXOs: exosomes; NC: Negative Control.
Further, results showed that exosome-induced migration (P < 0.001) and tube formation in TAECs, along with the inhibition of α-SMA expression, can be abolished by exosomes from inhibitor-treated lung cancer cells (P < 0.001) and partially abolished by exosome uptake inhibitors [Figure 7c-e]. Specifically, exosome-treated TAECs exhibited diminished E-cadherin, claudin-5, occludin, and Zonula Occludens-1(ZO-1) expression levels, and elevated N-cadherin and vimentin expression can be reversed through incubation with exosomes from inhibitor-treated lung cancer cells or through cytochalasin D treatment [Figure S2].
miR-5703 directly targets ING4
The targets of miR-5703 were predicted using the TargetScan database (https://www.targetscan.org/vert_80/). Using the WEB-Based Gene SeT AnaLysis Toolkit (https://www.webgestalt.org/), we found that the target gene set was associated with angiogenesis and lung cancer [Figure S3]. After the target set (cumulative weighted context++ score < −0.6, Total context++ score < −0.8) was screened, potential miR-5703 targets were obtained [Table S2]. Subsequently, the expression of each gene and prognosis was evaluated using the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn/index.html). ING4 was determined as the target of miR-5703 for subsequent experiments [Figures S4, S5, and 8a].
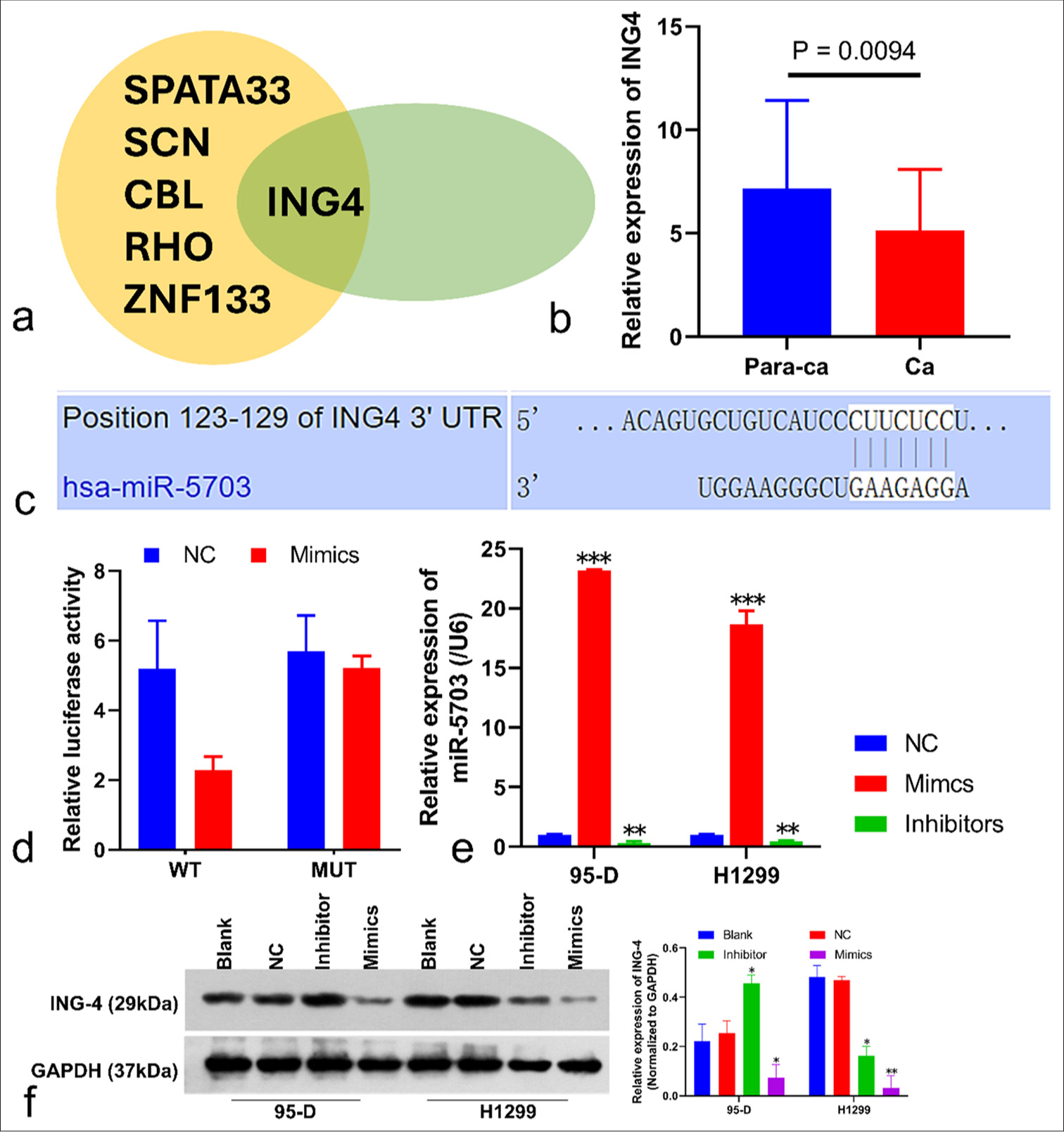
- Identification of the target genes of microRNA-5703 (miR-5703) and correlation analysis. (a) Inhibitor of growth family, member 4 (ING4) was screened as the target of miR-5703. SPATA: Spermatogenesis-Associated Transcript; SCN: Sodium Channel; CBL: Casitas B-lineage Lymphoma; RHO: Rhodopsin; ZNF133: Zinc Finger Protein 133. (b) The expression feature of ING4 in the clinical tissues of patients with lung cancer. (c) Binding site information for miR-5703 and ING4 as provided by the TargetScan database. (d) Verification of the relationship between miR-5703 and ING4 in 293T cells. WT: Wild Type; MUT: Mutant. (e and f) miR-5703-mediated suppression of ING4 expression observed on miR-5703 messenger RNA (e) and ING4 protein (f) levels. n = 3, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. NC: Negative Control; Para-ca: adjacent tissues; Ca: Lung cancer tissues.
The difference in ING4 expression between lung cancer and adjacent tissues was detected through qPCR [Figure 8b]. ING4 expression in cancer tissues was significantly lower than in the adjacent tissues (P = 0.0094), [Figure 8b]. The TargetScan database provided binding site information for miR-5703 and ING4 [Figure 8c]. Then, whether miR-5703 can directly interact with ING4 in 293T cells, which was determined with dual-luciferase reporter assays. miR-5703 overexpression significantly reduced luciferase activity [Figure 8d], and the transfection of miR-5703 mimic mediated the suppression of ING4 protein expression [Figure 8e and f].
Modulation of the biological function of TAECs by miR- 5703 through ING4
To determine the biological effects of miR-5703 in endothelial cells and explore the latent mechanism of miR-5703/ING4 interactions, we overexpressed miR-5703 and reinstated ING4 expression. As corroborated by the immunofluorescence and Western Blot results, mimic treatment subdued ING4 expression. Concurrently, after transfection with the ING4 overexpression vector, rapid increase in the expression of ING4 was observed [Figure 9a and b].
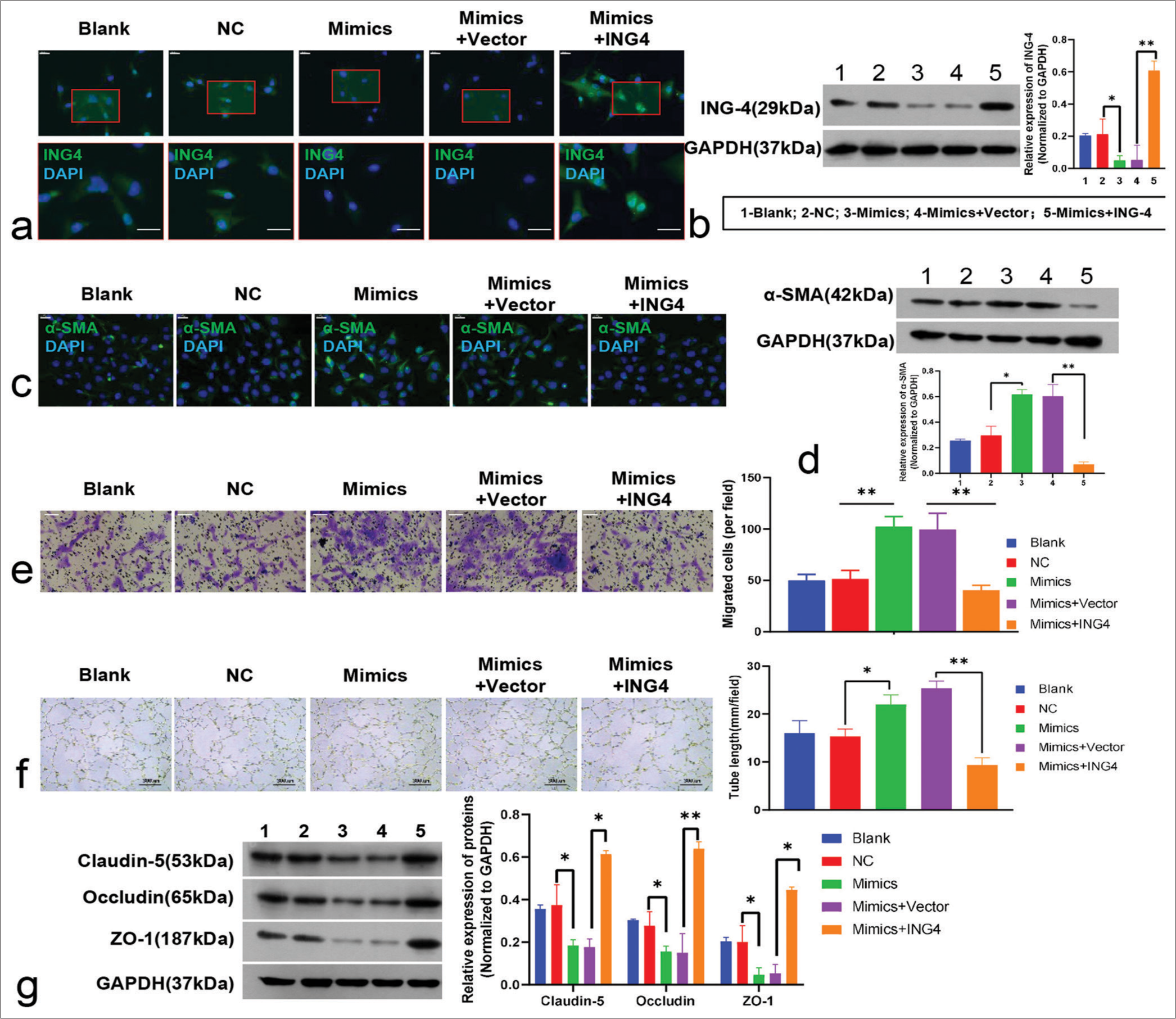
- Biological role of microRNA-5703 (miR-5703) in endothelial cells and functional investigation of miR-5703/Inhibitor of growth family, member 4 (ING4) interactions. (a and b) Results from immunofluorescence (a, scale: 20 μm) and Western blot (b) showing the effect of mimic treatment on ING4 expression. After transfection with the ING4 overexpression vector, an increase in ING4 expression was observed. (c and d) Immunofluorescence (c, Scale: 20 μm) and Western blot (d) illustrating increased α-smooth muscle actin levels in the endothelial cells after mimic treatment and reduced level after ING4 overexpression. (e) Transwell experiments displaying an increase in endothelial cell migration after mimic treatment and the reversal of this effect by ING4 overexpression. Scale: 50 μm. (f) Increased angiogenesis (blood vessel formation) in endothelial cells after mimic treatment and the reversal of this effect by ING4 overexpression. (g) Western blot showing the reduced expression of claudin-5, occludin, and ZO-1 after mimic treatment and restoration of their expression after ING4 overexpression. n = 3, ✶P < 0.05, ✶✶P < 0.01. NC: Negative Control; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; α-SMA: α-smooth muscle actin; ZO-1: Zonula Occludens-1.
The immunofluorescence and Western blot results also showed a heightened level of α-SMA in the endothelial cells treated with the mimics. By contrast, the overexpression of ING4 led to a reduction in the expression of α-SMA [Figure 9c and d]. In addition, as depicted in Figure 9e, the mimics promoted TAEC migration (P < 0.01) and tube formation and inhibited cell junctions, resulting in decreased expression of claudin-5, occludin, and ZO-1. However, the overexpression of ING4 reversed this mimics-induced effect (P < 0.01, [Figure 9e-g]).
miR-5703-loaded exosomes enhanced tumor growth and tumor microvasculature and impaired endothelial cell barrier function through ING4
To investigate the effect of exosomes encapsulated with miR-5703 on tumor growth in vivo, exosomes derived from lung cancer cells were administrated to animals, with miR-5703 inhibitors injected intratumorally as a remedial measure. As demonstrated in Figure 10, exosomes from lung cancer cells promoted tumor volume and mass growth (P < 0.05, [Figures 10a-d]). However, treatment with miR-5703 inhibitors significantly inhibited exosome-induced enhancement in tumor mass and volume (P < 0.01, [Figure 10a-d]). H&E staining revealed no significant histological changes in the tumor tissues after exosome treatment, but miR-5703 inhibitors led to areas of apoptosis [Figure 10e].
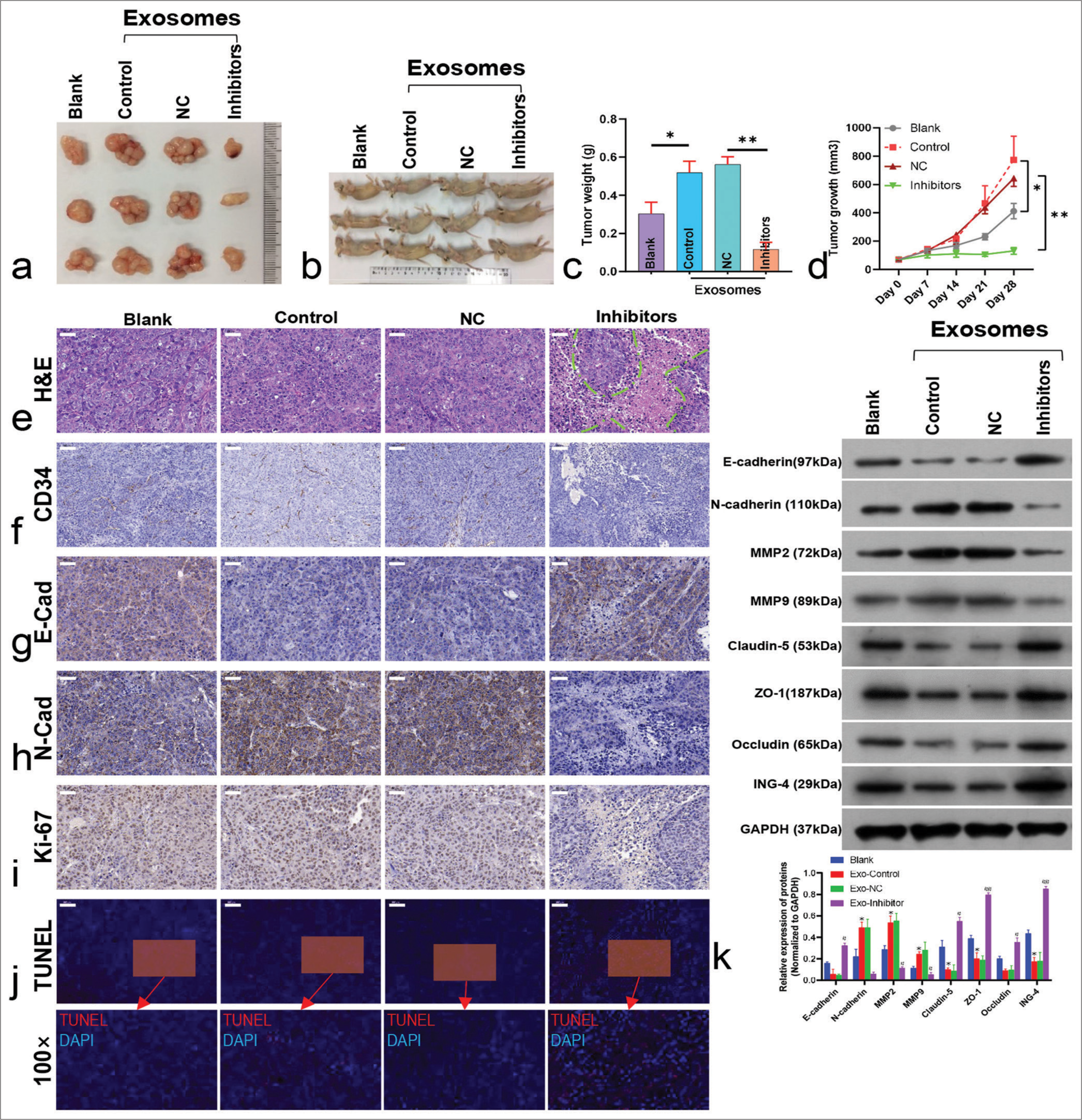
- Exosomal microRNA-5703 (miR-5703) and its inhibition affect tumor growth and protein expression. (a-d) Exosome treatment increased tumor volume and mass, but this effect was reversed by miR-5703 inhibitors. ✶P < 0.05, ✶✶P < 0.01. (e) Hematoxylin and eosin staining (H&E) showed apoptotic regions after miR-5703 inhibitor treatment. Green dotted lines highlighted the areas of tissue apoptosis and necrosis. (f-i) Immunohistochemical analysis revealed the altered expression of Cluster of Differentiation 34 (CD34) (f), N-cadherin (N-Cad, g), E-cadherin (E-Cad, h), and Kiloelectron Volts 67 (Ki-67). (i), in response to treatments. (j) Terminal deoxynucleotidyl transferase dUTP nick end labeling assay (TUNEL) indicated enhanced apoptosis with miR-5703 inhibitors. (k) Western blot analysis demonstrated changes in the expression of claudin-5, Zonula Occludens-1 (ZO-1), occludin, and Inhibitor of growth family, member 4 due to exosome and inhibitor treatments. MMP2: Matrix Metalloproteinase-2; MMP9: Matrix Metalloproteinase-9; ZO-1: Zonula Occludens-1; ING4: INhibitor of Growth 4; GADPH: glyceraldehyde-3-phosphate dehydrogenase. Scale: 50 μm. n = 5, ✶P < 0.05, ✶✶P < 0.01 versus Blank; #P < 0.05, ##P < 0.01 versus Exo-negative control. NC: Negative Control.
In addition, the expression of CD34, N-cadherin, and Ki-67 increased in the tumor tissues of the exosome-injected groups (including the inhibitor NC group) compared with the blank group, whereas E-cadherin expression decreased. Treatment with miR-5703 inhibitors significantly reduced the expression of CD34, N-cadherin, and Ki-67, while up-regulating E-cadherin [Figures 10f-i and S6a-d]. Moreover, exosome treatment did not alter TUNEL signaling in the tissues, which was enhanced by miR-5703 inhibitor treatment [Figure 10j]. Similarly, the results of Western blot analysis confirmed the immunohistochemistry findings for E-cadherin and N-cadherin expression. Furthermore, it was observed that exosome treatment decreased the expression of the intercellular junction proteins claudin-5, ZO-1, occludin, and ING4, while miR-5703 inhibitor treatment increased their expression levels [Figure 10k]. H&E staining showed that the cell nuclei of the adjacent noncancerous tissues were of the same shape, the nuclear staining was uniform and regular, and there were no significant pathological changes, while the cell atypia of the cancerous tissues was obvious, the tissue structure was disordered, and the growth pattern was invasive [Figure S7a]. The expression of CD34 and ING4 was determined using immunohistochemistry. CD34 was more extensively expressed in the cancerous tissues than in adjacent noncancerous tissues, whereas ING4 expression was down-regulated in the cancerous tissues [Figure S7b, c].
DISCUSSION
Lung cancer is the most prevalent malignant cancer worldwide.[24] China has a large smoking population, having high rates of morbidity and mortality due to lung cancer.[25] It is gradually emerging as a pivotal public health problem. Lung cancer has risen from the seventh leading cause of death in 1990 to the fourth leading cause of death by 2019.[26] How to alleviate this burden is an important issue related to national health and lifestyle.[27] Exosomes have attracted considerable interest in recent years. They have a mediating function in the development and advancement of lung cancer,[28] serving as drug carriers and possessing potential therapeutic effects.[29] This research mainly explored the mechanism of action of lung cancer cell-derived exosomal miR-5703 in the development of lung cancer.
Exosomes are minuscule extracellular nanovesicles that considerably contribute to cell–cell communication by disseminating vesicle contents, such as RNAs and proteins, to different cells. Functionally, they act as important messengers, mediating a diverse range of biological processes.[30] At present, research on exosomes in lung cancer primarily focuses on two aspects. Exosomes can promote the development of lung cancer; hence, they can be utilized as diagnostic markers for lung cancer.[31] Researchers have discovered a significant up-regulation of circular RNA (CircVMP1) in exosomes released from cisplatin-resistant non-small-cell lung cancer cells.[30] On the other hand, exosomes can participate in the occurrence and development mechanism of lung cancer. For instance, Zhou et al. engineered exosomes that actively deliver miR-449a (miR-449a Exo); their results suggest that the proliferation of A549 cells is effectively hampered by miR-449a Exo and this effect is accompanied by apoptosis in vivo and in vitro.[32] In this study, we primarily scrutinized clinical tissues, selected miRNA associated with lung cancer from five miRNAs, and detected miRNA in serum exosomes to ascertain its expression characteristics. The preliminary results indicated a correlation between miR-5703 and lung cancer occurrence, and miR-5703 expression positively correlated with the expression in serum exosomes. Hence, miR-5703 exhibited potential value in lung cancer diagnosis.
More and more researches focus on the role of exosomes in the regulation of the tumor microenvironment.[33,34] Exosomes released by tumor-associated macrophages encapsulates miR-223, promoting the lung metastasis of advanced breast cancer.[35] Exosomal miR-5703 can promote tumor progression, and exosomes secreted by pancreatic stellate cells mediate the PI3K/Akt pathway in pancreatic cancer cells with miR-5703, promoting cell proliferation and facilitating tumor growth in vivo and in vitro.[19] During tumor development, TAECs often undergo abnormal changes that disrupt blood vessel structure and function, thereby promoting tumor growth, metastasis, and invasion.[11] However, research on TAECs in lung cancer remains limited. In this study, we found that TAECs assimilated miR-5703 from lung cancer cell-derived exosomes, enhancing their angiogenic capability, augmenting EndMT characteristics, and weakening intercellular tight junctions. Claudin-5, Occludin, and ZO-1 are all tight junctions that play key roles in the composition and function of cellular barriers. In our study, we found that in 95-D cells, after inhibiting the expression of miR-5703 in exosomes, the expression of Claudin-5, Occludin, and ZO-1 increases; in H1299 cells, after inhibiting the expression of miR-5703 in exosomes, the expression of Claudin-5 and Occludinb increases. ZO-1 expression decreased. We speculated that the difference in the expression of ZO-1 in the two types of cells may be due to the different response mechanisms of different cell lines to the inhibition of miR-5703, including the regulatory differences in gene expression, the specific response of signaling pathways, and the diversity of cell states. On the inhibition of exosome uptake with cytochalasin D, related cellular functions were reversed. This result indicated the involvement of lung cancer cell-derived exosomal miR-5703 in the regulation of endothelial cells. However, further exploration is warranted to understand how miR-5703 functions in endothelial cells.
ING4 is a member of the ING family, possibly playing critical roles in numerous cellular processes, including angiogenesis and Epithelial-Mesenchymal Transition (EMT).[36] Overall, ING4 plays a definite tumor-suppressive role in lung cancer. Wang et al. have found, the expression of ING4 in both the nucleus and cytoplasm is reduced in lung cancer and is associated with tumor grading.[37] This may be closely associated with the regulatory effects of ING4 on angiogenesis and EMT. ING4 is widely recognized for its inhibitory function on angiogenesis. It has been reported that ING4 can suppress the expression of vascular endothelial growth factors in human retinal endothelial cells and inhibit their angiogenic function.[38] Research on lung cancer has suggested that ING4 has an inhibitory effect on tumor microvascular density in tumor-bearing nude mouse models, and this conclusion consistent with the results of this study.[39] In addition, the low expression of ING4 in lung cancer may also be associated with its inhibition of tumor EMT. According to research, ING4 exhibits significant inhibitory effects on the proliferation of Human Kidney 2 cells under hypoxic conditions, accompanied by upregulation of E-cadherin and downregulation of N-cadherin and vimentin expression.[40] In renal cell carcinoma, however, ING4 has a definite role in inhibiting EMT occurrence.[41] In this study, we found that ING4 has an inhibitory effect on the EndMT characteristics of endothelial cells after incubation with exosomes derived from lung cancer cells. TAECs typically exhibit high permeability, which implies a weakening of intercellular tight junctions.[11]
Although this study provides valuable insights, further research is necessary to explore miR-5703’s comprehensive effects and mechanistic pathways in the tumor microenvironment. Understanding its broader implications can facilitate the formulation of novel therapeutic strategies for lung cancer.
SUMMARY
This study focuses on elucidating three aspects: the promoting effects of miR-5703 on the malignant behavior of lung cancer cells, encapsulation and release of miR-5703 carried by exosomes by lung cancer cells, and TAECs uptaken the exosomes, leading to an increase in angiogenesis, EndMT and barrier dysfunction, ultimately providing a favorable condition for the occurrence and progression of lung cancer by tarting ING4. This study explains the involvement of miR- 5703/ING4 in lung cancer metastasis from the perspective of microenvironments and provides novel insights into the role of ING4 in angiogenesis.
ACKNOWLEDGMENT
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
ABBREVIATIONS
CCK8: Cell Counting Kit-8
DAPI: 4',6-diamidino-2'-phenylindole
EdU: 5-ethynyl-2'-deoxyuridine
EndMT: Endothelial–mesenchymal transition
FCM: Flow cytometry
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
H&E: Hematoxylin and eosin
ING4: Inhibitor of growth family member 4
miRNA: MicroRNA
NC: Negative control
PBS: Phosphate-Buffered Saline
PI: Propidium iodide
qRT-PCR: Quantitative real-time polymerase chain reaction
TAECs: Tumor-associated endothelial cells
TUNEL: Terminal deoxynucleotidyl transferase dUTP nick end labeling
α-SMA: α-smooth muscle actin
AUTHOR CONTRIBUTIONS
BW and ZFZ: Designed and performed research, analyzed data, and wrote manuscript; RCT and YYL: Assisted in experimental conception and design, oversaw experimental performance. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Second People’s Hospital of Yibin Ethics Committee, (No. 2023-162-01), August 2023. All experiments involving animals were approved by The Animal Ethical and Welfare Committee of Tianjin Medical University Cancer Institute and Hospital, (No. 2023078), September 2023. Informed consent was obtained from all study participants complies with the Declaration of Helsinki.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
This work was supported by Sichuan Provincial Medical Research Project Plan (No. S23026).
References
- Cancer progress and priorities: Lung cancer. Cancer Epidemiol Biomarkers Prev. 2019;28:1563-79.
- [CrossRef] [PubMed] [Google Scholar]
- Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623-40.
- [CrossRef] [PubMed] [Google Scholar]
- Anti-cancer effects of auranofin in human lung cancer cells by increasing intracellular ROS levels and depleting GSH levels. Molecules. 2022;27:5207.
- [CrossRef] [PubMed] [Google Scholar]
- Influential factors on treatment decision making among patients with colorectal cancer: A scoping review. Support Care Cancer. 2017;25:2943-51.
- [CrossRef] [PubMed] [Google Scholar]
- Drug exposure in a metastatic human lung adenocarcinoma cell line gives rise to cells with differing adhesion, proliferation, and gene expression: Implications for cancer chemotherapy. Mol Med Rep. 2015;12:3236-42.
- [CrossRef] [PubMed] [Google Scholar]
- BTBD7 downregulates E-cadherin and promotes epithelial-mesenchymal transition in lung cancer. Biomed Res Int. 2019;2019:5937635.
- [CrossRef] [PubMed] [Google Scholar]
- Vascular endothelial cell specification in health and disease. Angiogenesis. 2021;24:213-36.
- [CrossRef] [PubMed] [Google Scholar]
- Inhibition of endothelial FAK activity prevents tumor metastasis by enhancing barrier function. J Cell Biol. 2014;204:247-63.
- [CrossRef] [PubMed] [Google Scholar]
- NEO212, a conjugate of temozolomide and perillyl alcohol, blocks the endothelial-to-mesenchymal transition in tumor-associated brain endothelial cells in glioblastoma. Cancer Lett. 2019;442:170-80.
- [CrossRef] [PubMed] [Google Scholar]
- The dark side of tumor-associated endothelial cells. Semin Immunol. 2018;35:35-47.
- [CrossRef] [PubMed] [Google Scholar]
- Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther. 2018;188:1-11.
- [CrossRef] [PubMed] [Google Scholar]
- Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085-99.
- [CrossRef] [PubMed] [Google Scholar]
- Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93-105.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J Neurosurg. 2018;128:1091-101.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int J Oncol. 2012;40:130-8.
- [Google Scholar]
- Pancreatic stellate cells derived exosomal miR-5703 promotes pancreatic cancer by downregulating CMTM4 and activating PI3K/Akt pathway. Cancer Lett. 2020;490:20-30.
- [CrossRef] [PubMed] [Google Scholar]
- Bovine lactoferrin suppresses tumor angiogenesis through NF-κB Pathway inhibition by binding to TRAF6. Pharmaceutics. 2023;15:165.
- [CrossRef] [PubMed] [Google Scholar]
- Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:220677.
- [CrossRef] [PubMed] [Google Scholar]
- A review on PCR and POC-PCR-A boon in the diagnosis of COVID-19. Curr Pharm Anal. 2022;18:745-64.
- [CrossRef] [Google Scholar]
- Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of clinical responses to PD-1/PD-L1 inhibitors in non-small cell lung cancer: Predictive value of multidimensional immunomarker detection for the efficacy of PD-1 inhibitors in Chinese patients. Thorac Cancer. 2019;10:1303-9.
- [CrossRef] [PubMed] [Google Scholar]
- Attributable causes of lung cancer incidence and mortality in China. Thorac Cancer. 2011;2:156-63.
- [CrossRef] [PubMed] [Google Scholar]
- Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-53.
- [CrossRef] [PubMed] [Google Scholar]
- Burden of lung cancer in China, 1990-2019: Findings from the global burden of disease study 2019. Cancer Control. 2023;30:1-9.
- [CrossRef] [PubMed] [Google Scholar]
- Tumor cells-derived exosomal PD-L1 promotes the growth and invasion of lung cancer cells in vitro via mediating macrophages M2 polarization. Eur J Histochem. 2023;67:3784.
- [CrossRef] [PubMed] [Google Scholar]
- Bleomycin loaded exosomes enhanced antitumor therapeutic efficacy and reduced toxicity. Life Sci. 2023;330:121977.
- [CrossRef] [PubMed] [Google Scholar]
- Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022;29:1257-71.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNAs modulation in lung cancer: Exploring dual mechanisms and clinical prospects. Biocell. 2024;48:403-13.
- [CrossRef] [Google Scholar]
- Engineered exosomes loaded with miR-449a selectively inhibit the growth of homologous non-small cell lung cancer. Cancer Cell Int. 2021;21:485.
- [CrossRef] [PubMed] [Google Scholar]
- Pancreatic cancer-targeting exosomes for enhancing immunotherapy and reprogramming tumor microenvironment. Biomaterials. 2021;268:120546.
- [CrossRef] [PubMed] [Google Scholar]
- Tumor-derived exosomes elicit cancer-associated fibroblasts shaping inflammatory tumor microenvironment in head and neck squamous cell carcinoma. Oral Oncol. 2023;136:106270.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomal miRNA-223-3p derived from tumor associated macrophages promotes pulmonary metastasis of breast cancer 4T1 cells. Transl Oncol. 2023;35:101715.
- [CrossRef] [PubMed] [Google Scholar]
- miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55-65.
- [CrossRef] [PubMed] [Google Scholar]
- Down-regulation of ING4 is associated with initiation and progression of lung cancer. Histopathology. 2010;57:271-81.
- [CrossRef] [PubMed] [Google Scholar]
- Inhibitor of growth 4 affects hypoxia-induced migration and angiogenesis regulation in retinal pigment epithelial cells. J Cell Physiol. 2019;234:15243-56.
- [CrossRef] [PubMed] [Google Scholar]
- Enhanced tumor suppression by an ING4/IL-24 bicistronic adenovirus-mediated gene cotransfer in human non-small cell lung cancer cells. Cancer Gene Ther. 2011;18:627-36.
- [CrossRef] [PubMed] [Google Scholar]
- Inhibitor of growth 4 (ING4) inhibits hypoxia-induced EMT by decreasing HIF-1α and snail in HK2 cells. Acta Histochem. 2019;121:695-703.
- [CrossRef] [PubMed] [Google Scholar]
- Inhibitor of growth 4 inhibits cell proliferation, migration, and induces apoptosis of renal cell carcinoma cells. J Cell Biochem. 2019;120:6709-17.
- [CrossRef] [PubMed] [Google Scholar]








