Translate this page into:
Mechanistic insights into PDZK1-interacting protein 1 on the malignant progression of colorectal carcinoma

Chang Zhang

Leilei Hao
*Corresponding authors: Chang Zhang, Anorectal, The Affiliated Yantai Yuhuangding Hospital of Qingdao University, Yantai, Shandong Province, China. zhangchang3366@126.com
Leilei Hao, Anorectal, The Affiliated Yantai Yuhuangding Hospital of Qingdao University, Yantai, Shandong Province, China. qianer0904@163.com
-
Received: ,
Accepted: ,
How to cite this article: Yu K, Yu Y, Zhang C, Hao L. Mechanistic insights into PDZK1-interacting protein 1 on the malignant progression of colorectal carcinoma. CytoJournal. 2025;22:52. doi: 10.25259/Cytojournal_200_2024
Abstract
Objective:
PDZ domain containing 1-interacting protein 1 (PDZK1IP1) is commonly overexpressed in a wide variety of cancer. Hence, the objective of the present study is to ascertain the influences of PDZK1IP1 on colorectal carcinoma (CRC) development.
Material and Methods:
PDZK1IP1 expression was tested through reverse transcription-quantitative polymerase chain reaction and Western blot analysis, and its correlation with prognosis was analyzed using the GEPIA website. Small interfering RNA against PDZK1IP1 was adopted to downregulate PDZK1IP1 expression in CRC cells. The effects of PDZK1IP1 on cell growth were ascertained using colony formation and CCK-8 tests, and CRC cell apoptosis was analyzed through flow cytometry. Cell migration capability and invasiveness were measured using Matrigel Transwell and scratch-healing assays.
Results:
PDZK1IP1 was highly expressed in the CRC tissues (P < 0.001) and cells (P < 0.05), and its knockdown restrained cell growth (P < 0.05), migratory potential (P < 0.01), and invasive capacities (P < 0.001) and accelerated cell apoptosis (P < 0.001). Mechanically, PDZK1IP1 silencing blocked CRC progression by inactivating the phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of the rapamycin pathway.
Conclusion:
PDZK1IP1 contributes to the oncogenesis of CRC. This finding provides a basis for the diagnosis, treatment, and prevention of CRC.
Keywords
Colorectal carcinoma
PDZ domain containing 1-interacting protein 1
Phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin
INTRODUCTION
Colorectal carcinoma (CRC) is a common cancer that typically originates from intestinal mucosal epithelial cells and is one of the leading deadly cancer types worldwide. According to GLOBOCAN, CRC has the third highest global incidence (10.0%) after breast and lung cancer and the second highest mortality (9.4%),[1] posing a huge burden on human health.[2] Although the comprehensive application of various treatment strategies has improved the overall survival of patients with CRC,[3,4] clinical prognosis remains poor due to the high incidence of recurrence and metastasis in patients with advanced CRC. Therefore, searching for targets that contribute to the early diagnosis and treatment of the disease is of great significance to the survival of patients with CRC.
PDZ domain containing 1-interacting protein 1 (PDZK1IP1; commonly referred to as MAP17) is mainly found in the Golgi apparatus and plasma membrane.[5] PDZK1IP1 is only expressed in specific normal epithelial cells, including the spermatozoa of fine seminiferous tubules, epidermal keratinocytes, and renal proximal tubular epithelial cells, and is initially identified as an epigenetic-specific molecule.[6] Notably, PDZK1IP1 is commonly overexpressed in a wide variety of cancers[7] and is related to the malignancy of various tumors, and MAP17 promotes horizontal growth and movement by transferring neoplastic cells in the breast.[8] A previous study also found that PDZK1IP1 was an oncogene that correlated with tumorigenicity of papillary thyroid carcinoma (PTC).[6,9] It was reported that PDZK1IP1 is essential for the advancement of oral squamous cell carcinoma and gastric cancer by acting as a target for miR-455-5p and exosomal LINC00853.[10,11] Notably, high amounts of PDZK1IP1 have been detected in CRC tissues. Rivero et al. reported that high levels of pH2AX and MAP17 indicated the enhanced response of rectal cancer cells to radiotherapy and chemotherapy.[12] High MAP17 expression is an important risk factor for CRC morbidity and mortality in patients with early CRC.[13] However, the mechanism underlying the involvement of PDZK1IP1 in the occurrence and development of CRC is rarely reported.
The phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of the rapamycin (PI3K/AKT/mTOR) pathway participated in regulating cellular events, including promoting tumor growth, metabolism, apoptosis, proliferation, and metastasis.[14-16] To date, numerous studies have confirmed that this pathway exhibits a crucial role in CRC.[17,18] Zhu et al. reported that NUCKS1 regulates the development of CRC through PI3K/AKT/mTOR pathway.[19] Previous evidence revealed that LINC00853 can exert carcinogenic effects by binding to MAP17 and activating the PDZK1/AKT pathway in gastric cancer.[11] However, whether the effects induced by PDZK1IP1 on CRC are mediated through the PI3K/AKT/mTOR pathway is unclear. Here, the effect of PDZK1IP1 downregulation on CRC was investigated. The aim was to provide data for finding an effective target gene therapy for the disease.
MATERIAL AND METHODS
Bioinformatics analysis
TIMER database (https://cistrome.shinyapps.io/timer) was used in observing the expression of PDZK1IP1 in tumors.
Clinical samples
Our clinical samples (including tumor and paracancerous tissue samples; n = 9) were obtained from patients with CRC treated at our hospital from May to December 2023. The inclusion criteria were as follows: (i) Meet the diagnosis of CRC; (ii) absence of contraindication related to surgery; and (iii) absence of treatment before admission. The following patients were excluded: (i) patients who did not cooperate with the follow-up visit and (ii) those with missing case data. During the coloproctectomy of patients with CRC, fresh samples were collected. All the patients offered written informed consent. The study was conformed to the provisions of the Declaration of Helsinki[20] and approved by the ethics committee of our hospital (approval no. 2024163). The tissues were kept at −80°C before their utilization.
Cell lines and transfection
CRC cells (SW-620, cat. no. CL-0225; DLD-1, cat. no. CL-0074; HCT-116, cat. no. CL-0096; Caco2, cat. no. CL-0050) were purchased from Pricella Biotechnology Co., Ltd. (Wuhan, Hubei, China), whereas normal human colonic epithelial cells (NCM460, cat. no. TCH-C450) were purchased from Haixing Biosciences (Suzhou, China). The cells were tested negative for the presence of mycoplasma and were identified by short tandem repeat without cross-contamination. They were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (Gibco) at 37°C and 5% carbon dioxide (CO2) and 1% penicillin-streptomycin solution (cat. no. 15140122; Gibco, Grand Island, NY, USA). When the density of the cultured cells reached 75–90%, passage culture or other experimental procedures were performed after digestion with pancreatic enzymes.
For the silencing of PDZK1IP1, small interfering RNAs (siRNAs) targeting PDZK1IP1 (si-PDZK1IP1-1, 5'-CAAUCGCCUUUGCUGUCAATT-3'; si-PDZK1IP1-2, 5'-GUCAUGACCAUUGGAAACATT-3') and control scrambled siRNA (si-NC, 5'-CAAUCGCCUU UGCUGUCA ATT-3') were synthesized by Ribobio (Guangzhou, China). Lipo6000 (Beyotime, Shanghai, China) was adopted for the transfection. After 24 h, the cells were collected for further experiments.
Colonyformation tests
One day after transfection, SW-620 and HCT-116 cells were collected and cultivated in six-well plates. Afterwards, the cells were cultured at 37℃ with 5% CO2 for 10 days, fixed with 4% paraformaldehyde (cat. no. P0099-3L; Beyotime, Shanghai, China) for 15 min, and stained with 0.1% crystal violet (cat. no. C0121; Beyotime, Shanghai, China) for 15 min. After soaking with PBS twice, air drying was carried out, followed by photography and counting.
CCK-8 assay
The CRC cells treated with si-PDZK1IP1 were digested with 0.25% trypsin (cat. no. P4201; Beyotime, Shanghai, China), followed by 1000 r/min ultrafast centrifugation. The cell precipitates were collected. After the fresh and complete culture medium were blown evenly, the SW-620 and HCT-116 cells were counted with the abalone counting plate and then inoculated into a 96-well plate. Four wells were used for each group. After transfection at different time points, 10 μL of CCK-8 reagent (cat. no. C0037; Beyotime, Shanghai, China) was added to the cells, which were then incubated for 2 h. The mixture was shaken for 15 min in a shaker (Odd Instrument Co., Ltd, Beijing, China). The absorbance values of each group and each hole were measured and recorded using a microplate reader (DR-3518G, Hiwell Diatek, Wuxi, China) at 450 nm wavelength.
Scratch healing assay
Once the cell confluence reached 80%, the transfected cells were replaced with a free medium for starvation culture for 12 h. A 200 μL spearhead was used and placed perpendicular to the culture plate, and cell debris was washed off with PBS (cat. no. C0221A; Beyotime, Shanghai, China). DMEM (cat. no. C0891; Beyotime, Shanghai, China) was added to each well, and then culturing was continued at 37℃ in an incubator with 5% CO2. The scratch width was photographed under an optical microscope (OLYMPUS BX53; Olympus, Japan) at 0 and 24 h.
Transwell assay
Briefly, a 24-well plate was coated with Matrigel (8 μm) and incubated for 30 min at 37°C. Transfected SW-620 and HCT-116 cells were gently resuspended in 200 μL of the medium, which was then carefully added to the upper chamber. The lower chamber was filled with DMEM. The cells on the apical surface of the chambers were cultured for 48 h and then carefully removed. The cells at the bottom were then fixed and stained with crystal violet for 30 min. The cells were observed under an optical microscope (OLYMPUS BX53; Olympus, Japan).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The cells were exposed to Trizol reagent (cat. no. 15596018CN; Invitrogen, Carlsbad, CA, USA) for total RNA isolation. Complementary DNA synthesis was carried out with a Prime Script reverse transcription kit (cat. no. K1691; Invitrogen). Then, we conducted RT-qPCR analysis, utilizing Hieff qPCR SYBR Green Master Mix (cat. no. 11202ES08; Yeasen Biotechnology, Shanghai, China). To assess PDZK1IP1 level, we employed the 2−△△Ct method, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serving as an internal control: GAPDH F: 5'-GGAGCGAGATCCCTCCAAAAT-3', R: 5'- GGCTGTTGTCATACTTCTCATGG-3'; PDZK1IP1 F: 5'-TGTGTTCCTGGTCCTTGTCG-3', R: 5'-CCTCCTCCTC TGGGATGTTC-3'.
Western blot (WB) analysis
Proteins were gently extracted from tissues or cells using radioimmunoprecipitation assay (RIPA) buffer and then quantified using a BCA protein assay kit (cat. no. P0009; Beyotime, Shanghai, China). Equal amounts of proteins underwent Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and then transferred to polyvinylidene fluoride membranes (cat. no. GVWP04700; Millipore, Bedford, MA, USA). After this procedure, 5% skim milk was employed to gently block the membrane for 2 h. We then placed the primary antibodies on the membranes and incubated the samples overnight at 4°C. After washing the next day, we added the secondary antibodies and subsequently incubated the samples for 1 h. The primary antibodies were against PDZK1IP1 (cat. no. 12518-1-AP; Proteintech, Manchester, UK; 1:1000), mTOR (cat. no. ET1608-5; 1:1000), PI3K (cat. no. EM1701-62; 1:1000), AKT (cat. no. EM40507; 1:1000), p-mTOR (cat. no. HA600094; 1:1000), p-PI3K (cat. no. HA721672; 1:1000), p-AKT (cat. no. ET1607-73; 1:1000), and GAPDH (cat. no. R1210-1; 1:1000), and the second antibodies (cat. no. NBI01H; 1:10,000) were obtained from Huabio (Hangzhou, China). After washing, the proteins were developed using an ECL reagent (Millipore) and analyzed using Image J (NIH, Bethesda, MD, USA) under an imaging system (Bio-Rad, Hercules, CA, USA). GAPDH was used for normalization.
Flow cytometry
SW-620 and HCT-116 (2 × 105 cells/well) with si-PDZK1IP1 transfection were inoculated into six-well plates. The cells were washed with precooled PBS and resuspended in 500 μL of annexin V-FITC/PI (#45115, CST, Massachusetts, USA) mixture buffer for 15 min. Then, the cells were subjected to flow cytometry (cat. no. BDFACSAriaII; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) for apoptosis analysis.
Statistical analysis
Data in this work were manifested as mean ± SD, which were analyzed with the Statistical Package for the Social Sciences (SPSS) (version 21.0; BM SPSS Statistics, Armonk, NY, USA). Student’s t-tests were employed for the analysis of differences between the two groups. Variations among multiple groups were gauged by applying a one-way analysis of variance followed by Tukey’s multiple comparisons test. P < 0.05 indicated statistical significance.
RESULTS
PDZK1IP1expression in CRC
To ascertain the importance of PDZK1IP1 across CRC progression, we used The Cancer Genome Atlas Program and Gene Expression Profiling Interactive Analysis databasesatabases in examining the difference in PDZK1IP1 expression in CRC. The data revealed notably higher PDZK1IP1 expression in colon adenocarcinoma tissues (P < 0.05) [Figure 1a]. RT-qPCR and WB illustrated an increased level of PDZK1IP1 in CRC tissues than in normal tissues (P < 0.01) [Figure 1b-d]. The above results indicated that PDZK1IP1 plays a role in CRC progression.
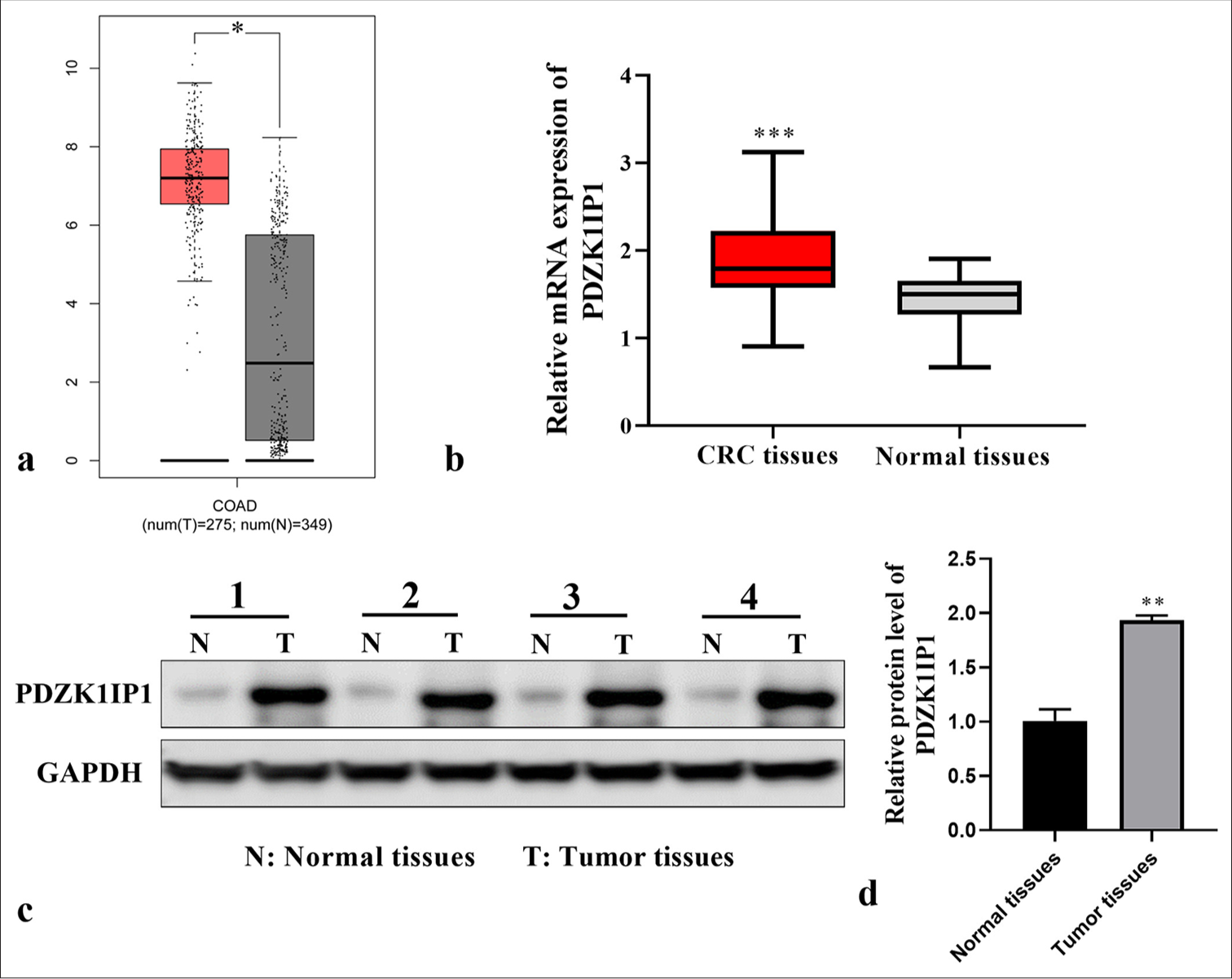
- PDZK1IP1 was upregulated in CRC. (a) PDZK1IP1 expression patterns in COAD and normal tissues were analyzed with the GEPIA databank. Red represents COAD tissues; black represents normal tissues. The vertical coordinate represents PDZK1IP1 messenger RNA expression. (b-d) PDZK1IP1 expression in CRC tissues was measured by RT-qPCR and WB. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. Normal tissues represent paracancerous tissue samples of patients with CRC. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, RT-qPCR: Reverse transcription-quantitative polymerase chain reaction, WB: Western blot, COAD: Colon adenocarcinoma, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
PDZK1IP1 facilitated the proliferative abilities and restrained the apoptosis of CRC cells in vitro
To clarify the effect of PDZK1IP1 on the advancement of CRC, RT-qPCR was performed on the CRC cells. A comparison between normal and CRC cells indicated that PDZK1IP1 was highly expressed in the CRC cells (SW-620, DLD-1, HCT-116, and Caco2, P < 0.05) [Figure 2a]. SW-620 and HCT-116 cells were transfected with si-PDZK1IP1, and the effectiveness of this procedure was validated by WB and RT-qPCR [Figure 2b-d] (P < 0.01). CCK-8 assay verified PDZK1IP1 lack restrained the viability of the CRC cells [Figure 2e and f] (P < 0.05). Colonyformation assay showed that silencing PDZK1IP1 subdued the colony numbers of the CRC cells [Figure 2g and h], (P < 0.01). Meanwhile, as shown in Figures 2i and j, after PDZK1IP1 silencing, the apoptosis rate of the CRC cells considerably increased (P < 0.001). Overall, these findings supported that PDZK1IP1performs a central role in CRC proliferation and apoptosis.
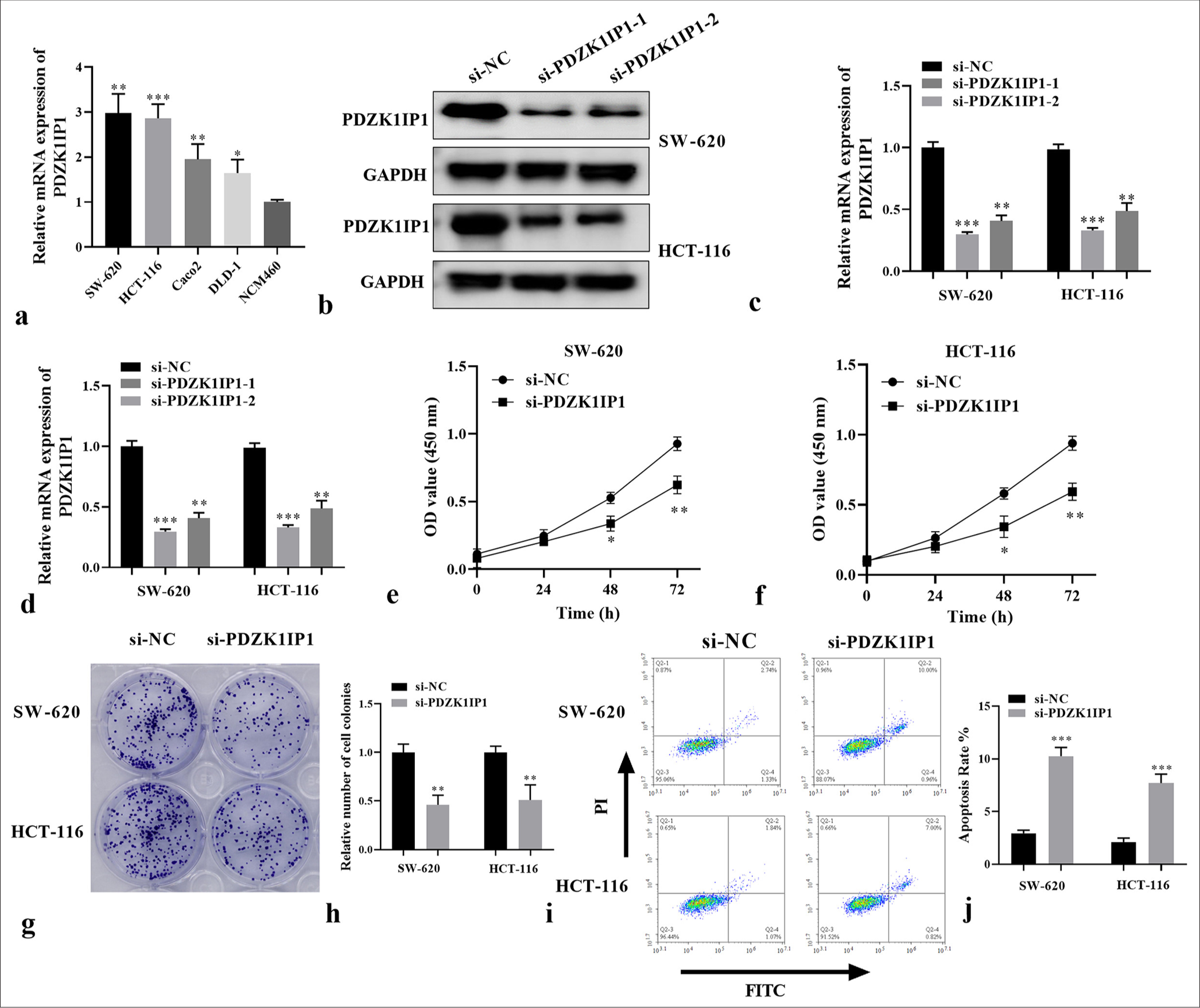
- PDZK1IP1 silencing restrained cell proliferation and promoted apoptosis in CRC. (a) The messenger RNA expression of PDZK1IP1 was examined by RT-qPCR in the CRC cells. (b-d) The protein expression of PDZK1IP1 was detected by Western blot and RT-qPCR after transfection. (e-h) The function of PDZK1IP1 on cell proliferation was evaluated by CCK-8 and colony formation assay. (i and j) Flow cytometric analysis of apoptosis in PDZK1IP1-knockdown SW-620 and HCT-116 cells. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, RT-qPCR: Reverse transcription-quantitative polymerase chain reaction, CCK-8: Cell counting kit-8, si-NC: Small interfering negative control, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, FITC, Fluorescein isothiocyanate.
Influences of PDZK1IP1 knockdown on CRC cell migration and invasion
PDZK1IP1’s role in cell migratory and invasive abilities was investigated in the CRC cells. Scratch healing assay showed that the migration activity of SW-620 and HCT-116 cells decreased in the si-PDZK1IP1 group versus the si-NC group (P < 0.01) [Figure 3a-d]. Invasion and migration ability changes in the transfected cells after PDZK1IP1 knockdown were investigated with the Transwell test. Migratory and invasive abilities were considerably reduced after PDZK1IP1 downregulation in the CRC cells (P < 0.001) [Figure 3e-h]. These data implied that the knockdown of PDZK1IP1 is associated with a decrease in cell migration and invasion in CRC.

- PDZK1IP1 knockdown suppresses the migration and invasion of CRC cells. (a-d) Cell migration of CRC cells with PDZK1IP1 knockdown was assessed by scratch healing assay. Scale bar = 200 μm, magnification × 40. (e-h) Analysis of cell invasion and migration of CRC cells with PDZK1IP1 knockdown was assessed by Transwell tests. Scale bar = 100 μm, magnification × 200. ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, si-NC: Small interfering negative control.
Influences of PDZK1IP1 knockdown on the PI3K/AKT/mTOR pathway
To elucidate the potential mechanism of PDZK1IP1 during CRC progression, we then ascertained the levels of proteins linked to the PI3K/AKT/mTOR pathway. The basal phosphorylation levels of PI3K, AKT, and mTOR dramatically decreased in the si-PDZK1IP1 group versus the control group in the SW-620 cells (P < 0.05) [Figure 4a-d]. These phosphorylated proteins markedly decreased in the HCT-116 cells with PDZK1IP1 silencing (P < 0.05). The findings indicated that PDZK1IP1 downregulation blocks this pathway in CRC cells.
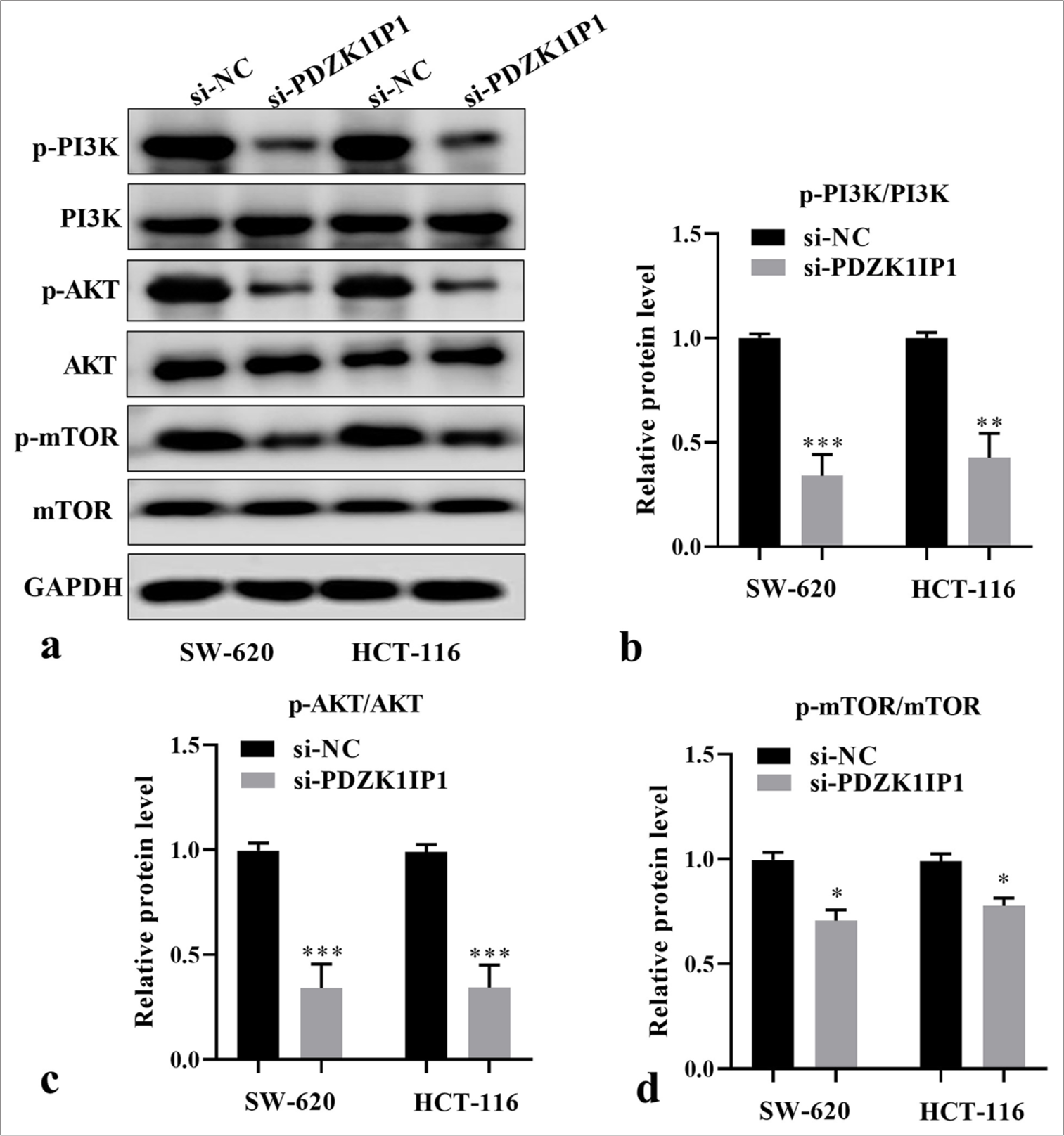
- PDZK1IP1 knockdown inhibited the PI3K/AKT/mTOR pathway of CRC cells. (a) Representative image of Western blot. (b-d) Relative protein levels of critical proteins in the PI3K/AKT/mTOR signaling pathway were detected by Western blot after PDZK1IP1 knockdown. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, PI3K/AKT/mTOR: Phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin, si-NC: Small interfering negative control, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, p-AKT: Phosphorylated Protein Kinase B, p-mTOR: Phosphorylated mechanistic target of rapamycin.
PDZK1IP1 regulates the malignant behaviors of CRC cells through PI3K/AKT/mTOR pathway
Next, the cellular functional experiments were performed to verify whether PDZK1IP1 regulates the malignant behavior of CRC through the PI3K/AKT/mTOR pathway. SC79, serving as a specific agonist of this pathway, was applied to activate this pathway. The CRC cells were transfected with si-PDZK1IP1 alone or with SC79. The results indicated that SC79 restored the phosphorylated protein levels decreased by PDZK1IP1 silencing (P < 0.05) [Figure 5a and b]. Furthermore, the decreased cell viability and proliferation rate triggered by si-PDZK1IP1 were eliminated by SC79 in the SW-620 cells (P < 0.05) [Figure 5c-e]. Concurrently, apoptosis considerably decreased in SC79-treated PDZK1IP1-silencing SW-620 cells compared with the untreated cells (P < 0.05) [Figure 5f and g]. Moreover, Figures 6a-e demonstrate that activating this pathway partially counteracted the influences of PDZK1IP1 knockdown on the migratory and invasive capacities of the SW-620 cells (P < 0.01). These results showed that PDZK1IP1 regulates CRC malignant behavior by activating this signaling pathway.

- PDZK1IP1 knockdown inhibited CRC cell growth and induced apoptosis by suppressing the PI3K/AKT/mTOR pathway. (a and b) The protein expression of critical proteins in the PI3K/AKT/mTOR signaling pathway was detected by Western blot. (c-e) The function of PDZK1IP1 and SC79 on cell proliferation was evaluated by CCK-8 and colony formation assay. (f and g) Flow cytometric analysis of apoptosis in PDZK1IP1-knockdown SW-620 cells after treatment with SC79. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, PI3K/AKT/mTOR: Phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin, CCK-8: Cell counting kit-8, si-NC: Small interfering negative control, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, p-AKT: Phosphorylated Protein Kinase B, p-mTOR: Phosphorylated mechanistic target of rapamycin, PI: Propidium iodide, FITC: Fluorescein isothiocyanate.
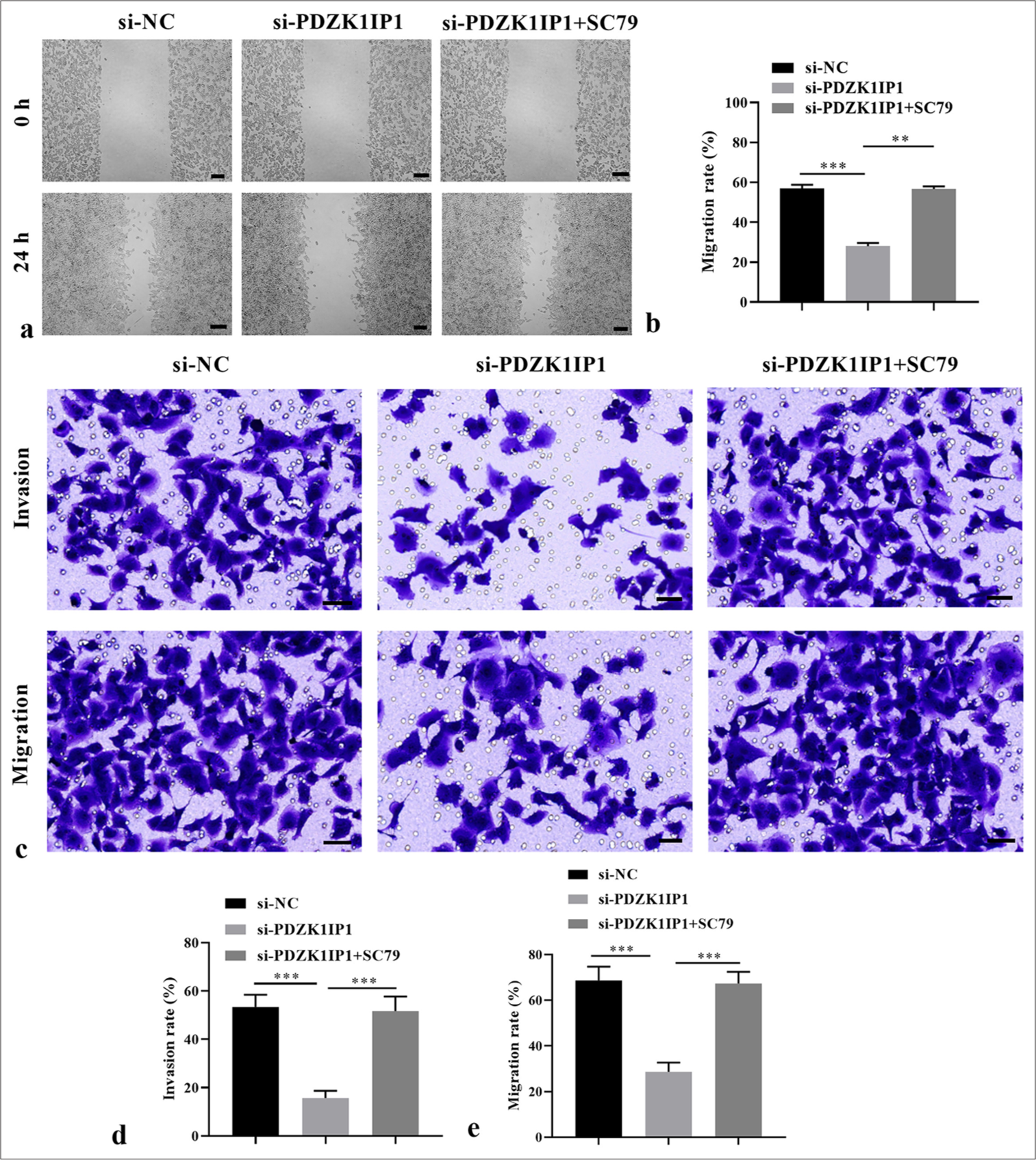
- PDZK1IP1 knockdown inhibited CRC cell migration and invasion by inhibiting the PI3K/AKT/mTOR pathway. (a and b) The function of PDZK1IP1 and SC79 on the migration of CRC cells was evaluated by scratch healing assay. Scale bar = 200 μm, magnification × 40. (c-e) The function of PDZK1IP1 and SC79 on migration and invasion of CRC cells was evaluated by Transwell assay. Scale bar = 100 μm, magnification × 200. ✶✶P < 0.01, ✶✶✶P < 0.001. Experiments were performed in triplicate, and each experiment was repeated 3 times. PDZK1IP1: PDZK1-interacting protein 1, CRC: Colorectal carcinoma, PI3K/AKT/mTOR: Phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin, si-NC: Small interfering negative control.
DISCUSSION
CRC is a common digestive tract tumor with increasing morbidity every year and affects young people.[21] Patients with CRC diagnosed at an early stage are 90% (5-year survival rate), compared to 13 % for those diagnosed at an advanced stage,[22] and about 35 % to 45 % of CRC patients with existing metastases relapse within 5 years of surgery.[23,24] Therefore, timely diagnosis and treatment are essential for enhancing the prognoses of patients with CRC. PDZK1IP1 is a prognostic factor for CRC,[12,13] but its effect on CRC remains largely unknown. We confirmed that PDZK1IP1 was highly expressed in CRC tissues and cells, and high PDZK1IP1 expression is associated with CRC progression. Functional experiments indicate that silencing PDZK1IP1 restrains the growth and movement of CRC cells. Moreover, we found that it can partially inactivate the PI3K/AKT/mTOR pathway and mediate CRC development.
Numerous literatures have proven that high PDZK1IP1 expression is linked to the tumorigenesis of various malignancies, including laryngeal, ovarian, and prostate cancer.[25-27] In addition, the overexpression of PDZK1IP1 is related not only to malignant characteristics and poor prognosis[8,28] but also to carcinogenic transformation and cell immortalization.[29] The upregulation of PDZK1IP1 elicited PTC cell proliferation, decreased apoptosis, and stimulated xenograft formation.[6] In line with the above research, bioinformatics analysis illustrated that PDZK1IP1 was upregulated in CRC tissues and cells. These results implied the oncogenic role of PDZK1IP1 during CRC progression. To further verify the effects of PDZK1IP1 on the malignant behavior of CRC, we silenced it in CRC cells with siRNA.
For the first time, we reported that the proliferation of the cells decreased, and apoptosis was enhanced after PDZK1IP1 silencing treatment in the CRC cells. Furthermore, after knocking down PDZK1IP1, cell migratory and invasive abilities were blocked. Overall, our study suggested that knocking down PDZK1IP1 suppressed the oncology behavior of the CRC cells.
The PI3K/AKT/mTOR pathway plays a considerable role in mediating CRC tumorigenesis.[16] Given that the important regulatory role of PDZK1IP1 in the development of CRC has been elucidated in the above results, we speculated that an interaction may exist between PDZK1IP1 and the PI3K/AKT/mTOR pathway. As expected, the present study uncovered that the phosphorylation levels of mTOR, PI3K, and AKT decreased when PDZK1IP1 was downregulated in the SW-620 and HCT-116 cells, implying that the PI3K/AKT/mTOR pathway can be inhibited by silencing PDZK1IP1. We then further speculated that PDZK1IP1 affects CRC’s oncology behavior by inducing the PI3K/AKT/mTOR pathway. The addition of SC79, which is a specific agonist of the PI3K/AKT/mTOR pathway, can remarkably counteract the PDZK1IP1’s effect that knocks down the malignant behavior of CRC cells. These results verified our hypothesis that PDZK1IP1 silencing mediates the inhibition of the PI3K/AKT/mTOR pathway and suppresses the tumorigenesis of CRC.
However, the present study has some limitations. First, we did not investigate how clinical features correlate with the expression of PDZK1IP1 in patients with CRC. Second, we did not perform in vivo experiments. Further experiments are needed to explore interactions between PDZK1IP1 and SC79 in CRC progression.
SUMMARY
The downregulation of PDZK1IP1 exerts tumor-suppressive effects in CRC, and this effect may be related to the PI3K/AKT/mTOR pathway. These results may provide some breakthrough for the treatment of CRC.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author on reasonable request.
ABBREVIATIONS
CCK-8: Cell counting kit-8
CRC: Colorectal carcinoma
PDZK1IP1: PDZK1-interacting protein 1
RT-qPCR: Reverse transcription-quantitative polymerase chain reaction
WB: Western blot
ACKNOWLEDGMENT
Not applicable.
AUTHOR CONTRIBUTIONS
KY.Y and YY: Conducted the research and contributed to data analysis and interpretation of the results; CZ and LL.H: Provided assistance and suggestions for the experiments. All authors participated in the drafting and critical revision of the manuscript. All authors have read and approved the final manuscript. All authors were fully involved in the work, able to take public responsibility for relevant portions of the content and agreed to be accountable for all aspects of the work, ensuring that any questions related to its accuracy or integrity are addressed. All authors meet ICMJE authorship requirements.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the Yantaishan Hospital (approval number 2024163). All patients provided written informed consent.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: Not applicable.
References
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-49.
- [CrossRef] [PubMed] [Google Scholar]
- Screening and prevention of colorectal cancer. BMJ. 2021;374:n1855.
- [CrossRef] [PubMed] [Google Scholar]
- The global challenge of colorectal cancer. Lancet Gastroenterol Hepatol. 2019;4:894-5.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA. 2021;325:669-85.
- [CrossRef] [PubMed] [Google Scholar]
- Pdzk1-interacting protein 1(pdzkip1) inhibits goat subcutaneous preadipocyte differentiation through promoting autophagy. Animals (Basel). 2023;13:1046.
- [CrossRef] [PubMed] [Google Scholar]
- Pdzk1 interacting protein 1 promotes the progression of papillary thyroid cancer. J Clin Endocrinol Metab. 2022;107:2449-61.
- [CrossRef] [PubMed] [Google Scholar]
- The expression and survival significance of sodium glucose transporters in pancreatic cancer. BMC Cancer. 2022;22:116.
- [CrossRef] [PubMed] [Google Scholar]
- Breast tumor cells promotes the horizontal propagation of EMT, stemness, and metastasis by transferring the map17 protein between subsets of neoplastic cells. Oncogenesis. 2020;9:96.
- [CrossRef] [PubMed] [Google Scholar]
- Pdzk1ip1 gene promotes proliferation, migration, and invasion in papillary thyroid carcinoma. Pathol Res Pract. 2022;238:154091.
- [CrossRef] [PubMed] [Google Scholar]
- Mir-455-5p suppresses pdzk1ip1 to promote the motility of oral squamous cell carcinoma and accelerate clinical cancer invasion by regulating partial epithelial-to-mesenchymal transition. J Exp Clin Cancer Res. 2023;42:40.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomal linc00853 promotes progression of gastric cancer via the map17/pdzk1/AKT signaling pathway. Noncoding RNA Res. 2024;9:876-86.
- [CrossRef] [PubMed] [Google Scholar]
- Map17 (pdzk1ip1) and ph2ax are potential predictive biomarkers for rectal cancer treatment efficacy. Oncotarget. 2018;9:32958-71.
- [CrossRef] [PubMed] [Google Scholar]
- Map17 expression in colorectal cancer is a prognostic factor for disease recurrence and dismal prognosis already in early stage disease. Oncology. 2021;99:471-82.
- [CrossRef] [PubMed] [Google Scholar]
- Attacking the pi3k/AKT/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin Cancer Biol. 2022;85:69-94.
- [CrossRef] [PubMed] [Google Scholar]
- Pi3k/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138.
- [CrossRef] [PubMed] [Google Scholar]
- Pi3k/AKT/mTOR signaling pathway as a target for colorectal cancer treatment. Int J Mol Sci. 2024;25:3178.
- [CrossRef] [PubMed] [Google Scholar]
- The pi3k/AKT/mTOR axis in colorectal cancer: Oncogenic alterations, non-coding RNAs, therapeutic opportunities, and the emerging role of nanoparticles. J Cell Physiol. 2022;237:1720-52.
- [CrossRef] [PubMed] [Google Scholar]
- Curcumin represses colorectal cancer cell proliferation by triggering ferroptosis via pi3k/AKT/mTOR signaling. Nutr Cancer. 2023;75:726-33.
- [CrossRef] [PubMed] [Google Scholar]
- Nucks1 promotes the progression of colorectal cancer via activating pi3k/AKT/mTOR signaling pathway. Neoplasma. 2023;70:272-86.
- [CrossRef] [PubMed] [Google Scholar]
- World medical association declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2025;333:71-4.
- [CrossRef] [PubMed] [Google Scholar]
- Long noncoding RNA bbox1-as1 increased radiotherapy sensitivity in colorectal cancer by stabilizing and activating pfk1. Transl Oncol. 2023;36:101751.
- [CrossRef] [PubMed] [Google Scholar]
- Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-44.
- [CrossRef] [PubMed] [Google Scholar]
- Rp11-296e3.2 acts as an important molecular chaperone for ybx1 and promotes colorectal cancer proliferation and metastasis by activating stat3. J Transl Med. 2023;21:418.
- [CrossRef] [PubMed] [Google Scholar]
- Knockdown of ribosomal protein L22-like 1 arrests the cell cycle and promotes apoptosis in colorectal cancer. Cytojournal. 2024;21:45.
- [CrossRef] [PubMed] [Google Scholar]
- Fxr1 associates with and degrades pdzk1ip1 and atoh8 mRNAs and promotes esophageal cancer progression. Biol Direct. 2024;19:104.
- [CrossRef] [PubMed] [Google Scholar]
- Spatiotemporal transcriptomic changes of human ovarian aging and the regulatory role of foxp1. Nat Aging. 2024;4:527-45.
- [CrossRef] [PubMed] [Google Scholar]
- The molecular function of kallikrein-related peptidase 14 demonstrates a key modulatory role in advanced prostate cancer. Mol Oncol. 2020;14:105-28.
- [CrossRef] [PubMed] [Google Scholar]
- Dr. Jekyll Mr. Hyde: Map17's up-regulation, a crosspoint in cancer and inflammatory diseases. Mol Cancer. 2018;17:80.
- [CrossRef] [PubMed] [Google Scholar]
- 80map17 promotes the tumorigenesis of papillary thyroid carcinoma by reducing the stability of p53. Front Biosci (Landmark Ed). 2021;26:777-88.
- [CrossRef] [PubMed] [Google Scholar]








