Translate this page into:
Relationship between transmembrane emp24 domain containing 2 expression and tumor stem cell characteristics in oral cancer

*Corresponding author: Guimei Zou, Department of Stomatology, Nanjing Integrated Traditional Chinese and Western Medicine Hospital, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, Jiangsu Province, China. 15365105162@163.com
-
Received: ,
Accepted: ,
How to cite this article: Wu Y, Zhang S, Zou G. Relationship between transmembrane emp24 domain containing 2 expression and tumor stem cell characteristics in oral cancer. 2025;22:5. CytoJournal. doi: 10.25259/Cytojournal_132_2024
Abstract
Objective:
Transmembrane Emp24 Domain Containing 2 (TMED2) is a mediator of membrane protein trafficking involved in intracellular protein transport. Recent research suggests that TMED2 plays an important role in the development and metastasis of tumors; however, its exact mechanisms in oral cancer (OC) remain unclear. This study aims to elucidate the role and possible mechanisms of TMED2 in OC.
Material and Methods:
We investigated the impact of TMED2 knockdown on the invasion, migration, and proliferation capabilities of OC cells. Furthermore, we analyzed the in vitro and in vivo interactions between TMED2 and polypeptide-N-acetylgalactosaminyltransferase 7 (GALNT7) as well as explored the regulatory function of TMED2 on GALNT7. The alterations in stem cell markers were assessed using clone formation assays, western blot, and quantitative real-time polymerase chain reaction.
Results:
The upregulation of TMED2 promoted the proliferation and invasion abilities of OC cells. Further analysis revealed that TMED2 enhanced the stem-like properties and tumorigenicity of OC cells by directly regulating the expression of GALNT7. In vivo and in vitro results suggested that silencing TMED2 expression reduced the incidence of OC.
Conclusion:
Our data imply that TMED2 stimulates GALNT7 transcription, which in turn amplifies the stem-like characteristics and carcinogenic potential of OC cells. Moreover, the block of TMED2 prevents cancers from growing and spreading in vivo. This finding provides a new therapeutic target for the treatment of OC and highlights the critical role of TMED2 in the condition.
Keywords
Oral cancer
Polypeptide-N-acetyl-galactosaminyltransferase 7
Transmembrane emp24 domain containing 2
INTRODUCTION
Oral cancer (OC) is a common malignant tumor that significantly affects the quality of life and overall health of a patient.[1,2] The formation and progression of OC are considerably influenced by the tumor microenvironment and the properties of cancer stem cells (CSCs).[3-5] CSCs exhibit self-renewal, proliferation, invasion, and drug resistance capabilities, which contribute to treatment resistance and metastasis.[6,7] Thus, understanding the molecular processes that control the stem-like characteristics of OC cells is important for developing innovative treatment approaches.[8,9]
Transmembrane Emp24 Domain Containing 2 (TMED2) is crucial for controlling tumor growth.[10,11] TMED2 is a transmembrane protein involved in the transport process between the endoplasmic reticulum and the Golgi apparatus.[12]
Research has demonstrated that TMED2 enhances the capacity of several tumor cells to proliferate and invade, such as breast cancer, lung cancer, and stomach cancer.[11,13] TMED2 improves tumor cell proliferation and invasion by regulating multiple signaling pathways and related genes, such as phosphoinositide 3-kinase/protein kinase B, extracellular regulated protein kinases, and Wnt/β-catenin.[14] Furthermore, TMED2 is associated with stem cell characteristics in various tumors.[15,16] The upregulation of TMED2 not only enhances the stem cell traits, self-renewal, and proliferation of CSCs but also exerts its effects by regulating the expression and/or activity of polypeptide-Nacetylgalactosaminyltransferase 7 (GALNT7).[15,16] GALNT7 is an acyltransferase involved in glycosylation modifications and tumor occurrence,[16-18] but its specific functions and mechanisms in OC are still unclear. Therefore, this study aims to explore the role of TMED2 in ovarian cancer. It also seeks to reveal the relationship between TMED2 and GALNT7. The focus is on the regulating effect of TMED2 on the function and expression of GALNT7, which influences the development of ovarian cancer.
We first evaluated the function of TMED2 in vitro cell models. Then, we explored the effect of TMED2 on the invasion and migration capabilities of OC cells. Subsequent research revealed a strong connection between TMED2 and GALNT7 in OC. Our results showed that the upregulation of GALNT7 is closely associated with enhanced stem-like properties and tumorigenicity in OC, including self-renewal, proliferation, invasion, and drug resistance capabilities. Therefore, we hypothesize that TMED2 enhances the stem-like properties and tumorigenicity of OC cells by promoting the transcription of GALNT7.
Our study may offer new insights into the mechanisms of OC development and progression. Further, research will help uncover the specific functions and mechanisms of TMED2 and GALNT7 in OC, as well as their potential as biomarkers for prognosis assessment and treatment response monitoring.
MATERIAL AND METHODS
Cell culture
In the cell culture experiment, human normal oral epithelial cells (CP-H203, Pricella, Wuhan, Hubei, China) and OC cells including TSCCa (CL-0235, Pricella, Wuhan, Hubei, China) and Tca-8113 (CL-0230, Pricella, Wuhan, Hubei, China) were cultured in high glucose Dulbecco’s modified eagle medium (C3260, Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS) (S9030, Solarbio, Beijing, China). The cells were incubated in a humidified atmosphere with 5% CO2 at 37°C until they reached 80% confluency. At this point, they were collected for further analysis. All cells were authenticated using short tandem repeat profiling, and mycoplasma testing was performed to avoid contamination.
Cell transfection
The cells were trypsinized for 2 min and centrifuged for 15 min. Subsequently, 5 × 106 cells were seeded in a 6-well plate. When the fusion rate reached 70%, the cells were transfected with negative control to TMED2 shRNA (sh-NC), TMED2 shRNA (sh-TMED2) (forward: 5’-CCGGTTCTCCGAACGTGTC A C G T T T C A A G A G A A C G T G A C A C G TTCGGAGAATTTTTG-3’, reverse: 5’-AATTCAAA AATTCTCCGAACGTGTCACGTTCTCTTGAAAC GTGACACGTTCGGAGAA-3’), OE-vector, or OEGALNT7 (forward: 5’-GCAGTCTCCTTTTAGTA ACTGCAGATGATAATGT-3’, reverse: 5’-GGGGGC CGGCCGTAATCCCAA AGAAGACAA-3’) using Lipofectamine 2000 (12566014, Thermo Fisher Scientific, Waltham, MA, USA). The cells were cultivated for a further 24 h after the media were switched to serum-free medium after 12 h. Thereafter, the total RNA of the cells was removed, and the effectiveness of the transfection was confirmed.
Establishment of xenograft tumor models
A total of 30 male Blab/c nude mice (4–6 weeks) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (SCXK [Jing] 2016-0006). All mice were placed in a specific pathogen-free environment at 25 ± 2°C and a 12/12 h light/dark cycle. Three groups of mice were randomly selected and injected subcutaneously with Tca-8113 cells (6 llst6) into the right axilla: Control+sh-NC+OE-vector, Control+sh-NC+OE-GALNT7, and Control+shTMED2+OE-vector. The time of tumor formation was recorded, the long diameter (a) and short diameter (b) of the tumor were measured every 7 days, and the tumor volume was calculated using the formula V = ab2/2. All mice were killed by cervical dislocation on day 28 after inoculation with Tca-8113 cells. Xenograft tumors and lungs were collected. The collected tissues were randomly divided into two groups: one group was fixed in 4% paraformaldehyde and the other group was frozen in the refrigerator at −80℃. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Nanjing Integrated Traditional Chinese and Western Medicine Hospital, Affiliated Hospital of Nanjing University of Chinese Medicine, approval No. 2024050.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using the Trizol (R1100, Solarbi, Beijing, China) method, followed by DNase I treatment to eliminate genomic DNA contamination. The isolated RNA was then converted into complementary DNA (cDNA) through reverse transcription (RP1105, Solarbio, Beijing, China). PCR amplification was conducted to assess the expression of the target gene using SYBR Green reagent (A46012, Thermo Fisher Scientific, Waltham, MA, USA) with cDNA as the template and b-actin as the internal control. The 2-ΔΔCt technique was used to analyze the obtained experimental data. Table 1 provides an overview of the particular primer sequences.
| Primes name | Primer sequences (5'-3') |
|---|---|
| TMED2-F | AGCUAGAAGAAAUGAUCAAUG |
| TMED2-R | GGUUCAAGUCACUAAAGAUUC |
| MYC-F | CGGCAGCGATAGCATAAAAT |
| MYC-R | ACCTCGTCGGTAAGACTGTGA |
| GALNT7-F | GGTACCATGGCCTCATGTTG |
| GALNT7-R | GCCACCACACTGCCATATCT |
| SOX2-F | AACCCCAAGATGCACAACTC |
| SOX2-R | CGGGGCCGGTATTTATAATC |
| KRT18-F | CACAGTCTGCTGAGGTTGGA |
| KRT18-R | GAGCTGCTCCATCTGTAGGG |
| CD24-F | TAGATGCCCCCAAATCTCAG |
| CD24-R | CTCATGGAGTCCAGGTCGAT |
| Nanog-F | TTCCTTCCTCCATGGATCTG |
| Nanog-R | TCTGCTGGAGGCTGAGGTAT |
| BMI1-F | CCAGGGCTTTTCAAAAATGA |
| BMI1-R | CCGATCCAATCTGTTCTGGT |
| β-actin-F | GGGAAATCGTGCGTGACATTAAG |
| β-actin-R | TGTGTTGGCGTACAGGTCTTTG |
TMED2: Transmembrane Emp24 Domain Containing 2, GALNT7: Polypeptide-N-acetylgalactosaminyltransferase 7, SOX2: SRY-box transcription factor 2, KRT18: Keratin 18, MYC: Myelocytomatosis oncogene, BMI1: B-cell-specific Moloney murine leukemia virus insertion site 1, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Western blot
Total protein was extracted from each group after 48 h of cell culture using 100 μL of cell lysis buffer (R0010, Solarbio, Beijing, China) on ice, and its concentration was quantified with a bicinchoninic acid assay kit (PC0020, Solarbio, Beijing, China). Sodium dodecyl sulfate polyacrylamide gel electrophoresis was prepared, and equal amounts of denatured protein samples were loaded and electrophoresed. Thereafter, the proteins were transferred on a polyvinylidene fluoride membrane (IPVH00010, Millipore Corporation, MA, USA) using a wet transfer technique, blocked with 5% skim milk, and incubated overnight with TMED2 (dilution: 1:500, ab251705, Abcam, Cambridge, MA, USA), GALNT7 (dilution: 1:1000, ab254971, Abcam, Cambridge, MA, USA), SRY-box transcription factor 2 (SOX2, dilution:1:1000, ab92494, Abcam, Cambridge, MA, USA), Octamer-Binding Transcription Factor 4 (OCT4, dilution:1:1000, ab181557, Abcam, Cambridge, MA, USA), Nanog (dilution:1:1000, ab109250, Abcam, Cambridge, MA, USA), and glyceraldehyde-3-phosphate dehydrogenase (dilution: 1:2000, ab8245, Abcam, Cambridge, MA, USA). After the membrane was washed using (tris-buffered saline with Tween 20, it was incubated with horseradish peroxidase-conjugated secondary antibodies (dilution: 1:2000, ab6721, Abcam, Cambridge, MA, USA). Using an enhanced chemiluminescence (BL520B, Biosharp Life Science, Hefei, Anhui, China) detection reagent, the image was captured through a computer imaging system (Image Quant LAS4000, GE Healthcare, Chicago, IL, USA). The gray values of protein bands were quantitatively analyzed by Image J (v1.3.4, National Institutes of Health, Bethesda, MD, USA).
5-ethynyl-2’-deoxyuridine (EdU) assay
First, a EdU solution (Jiangsu Kaqi Biotechnology Co., Ltd., lot: KGA331, Jiangsu, Zhejiang, China) was used. Then, 1 × 105 cells were inoculated in a 96-well plate. The cells were fixed with 4% paraformaldehyde for 30 min. Thereafter, 100 μL penetrant (phosphate-buffered saline [PBS] of 0.5% TritonX-100) was added to each well, followed by incubation for 10 min. After the cells were cleaned with PBS, they were stained according to the kit instructions. Next, 4’,6-diamidino-2-phenylindole (D8200, Solarbio, Beijing, China) staining solution was added, followed by incubation in the dark for 20 min. Finally, the cells were observed and photographed using a fluorescence microscope (CX41-32RFL, Olympus Corporation, Tokyo, Japan).
Transwell migration assay
A total of 1 × 105 cells were inoculated in Transwell insertion lumen (Corning Costar, Cambridge, MA, USA) to evaluate the migration and invasion ability of the cells. In the invasion experiment, the upper cavity was coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). An 800 μL medium containing 20% FBS was added to the lower cavity, followed by incubation at 37℃ for 48 h. After the incubation period, the upper cavity was removed, the inside of the film was gently swabbed, and the membrane was washed twice. The cells were then stained with 5× crystal violet (C8470, Solarbio, Beijing, China), washed, and air-dried upside down. Finally, the results were observed and analyzed using a microscope (CKX41, Olympus Corporation, Tokyo, Japan).
Colony formation assay
A sterile culture dish was prepared for seeding 1 × 103 cells. The dish was gently agitated to ensure uniform cell distribution and incubated in an incubator. After 14 days of culture, the formation of cell colonies was assessed. The colonies were fixed and stained with 0.5% crystal violet (G1065, Solarbio, Beijing, China), rinsed with water, and air-dried. Number of cell colonies was counted, with clusters of ≥50 cells regarded as one colony.
Cell counting kit-8 (CCK-8)
The cells were cultured in 96-well plates at a density of 4 × 103 cells/well and incubated overnight. A 10 μL CCK-8 (CA1210, Solarbio, Beijing, China) solution was added, followed by incubation at 37℃ for 1.5 h. Then, absorbance was measured at 450 nm (SpectraMax 190, Molecular Devices, San Jose, CA, USA).
Histopathology and immunohistochemistry assay
The fixed tumor tissue and lung were embedded in paraffin wax, sliced into 5 μm sections, and stained by hematoxylin and eosin (H and E) staining. After dewaxing and hydration, the sections were stained with hematoxylin for 5 min. Then, they were differentiated and redyed with 0.5% eosin dye for 10 s. Finally, the tax refund was sealed with neutral gum. The reagents used in these steps were from H and E kits (G1120, Solarbio, Beijing, China). The metastases of lung foci were observed by microscope (CKX41, Olympus Corporation, Tokyo, Japan). The expression level of Ki67 (ab15580, Abcam, Cambridge, MA, USA) in tumor sections was determined by immunohistochemistry. After the sections were dewaxed and hydrated, the Ki67 antibody was incubated overnight with 5% bovine serum albumin (SW3015, Solarbio, Beijing, China) group at 4℃. The secondary antibody (SA00004-2, Proteintech, Hubei, China) was then incubated at 37℃ for 2 h. The presence of brown-yellow granules indicated positive expression.
Expression profiling of the cancer genome atlas (TCGA) dataset
Sangerbox 3.0 (http://vip.sangerbox.com/) was utilized to download and analyze TMED2 and GALNT7 expression data along with related clinical information from TCGA data.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA, USA). Data for normally distributed quantitative variables were presented as mean ± standard deviation. Multiple group comparisons were conducted using analysis of variance, followed by Bonferroni’s post-hoc test, with α = 0.05. Comparisons between the two groups were analyzed using an independent sample t-test. A significance level of P < 0.05 was considered statistically significant.
RESULTS
TMED2 promotes the proliferation of OC cells in vitro
The abundance of TMED2 in human oral epithelial cells (HOEC), Tca-8113, and TSCCa was measured by RT-qPCR. TMED2 expression was lowest in HOEC cells, while it was higher in OC cells [P < 0.01, Figure 1a]. The expression levels of TMED2 protein in OC cell lines with sh-NC and sh-TMED2 were also measured. Compared with that in the sh-NC group, the abundance of TMED2 protein in the sh-TMED2 group was lower [P < 0.01, Figure 1b and c]. Results from the EdU assay showed that the sh-NC group had a higher number of positive cells, which indicated higher proliferative activity; meanwhile, the sh-TMED2 group had a lower number of positive cells [P < 0.001, Figure 1d and e].
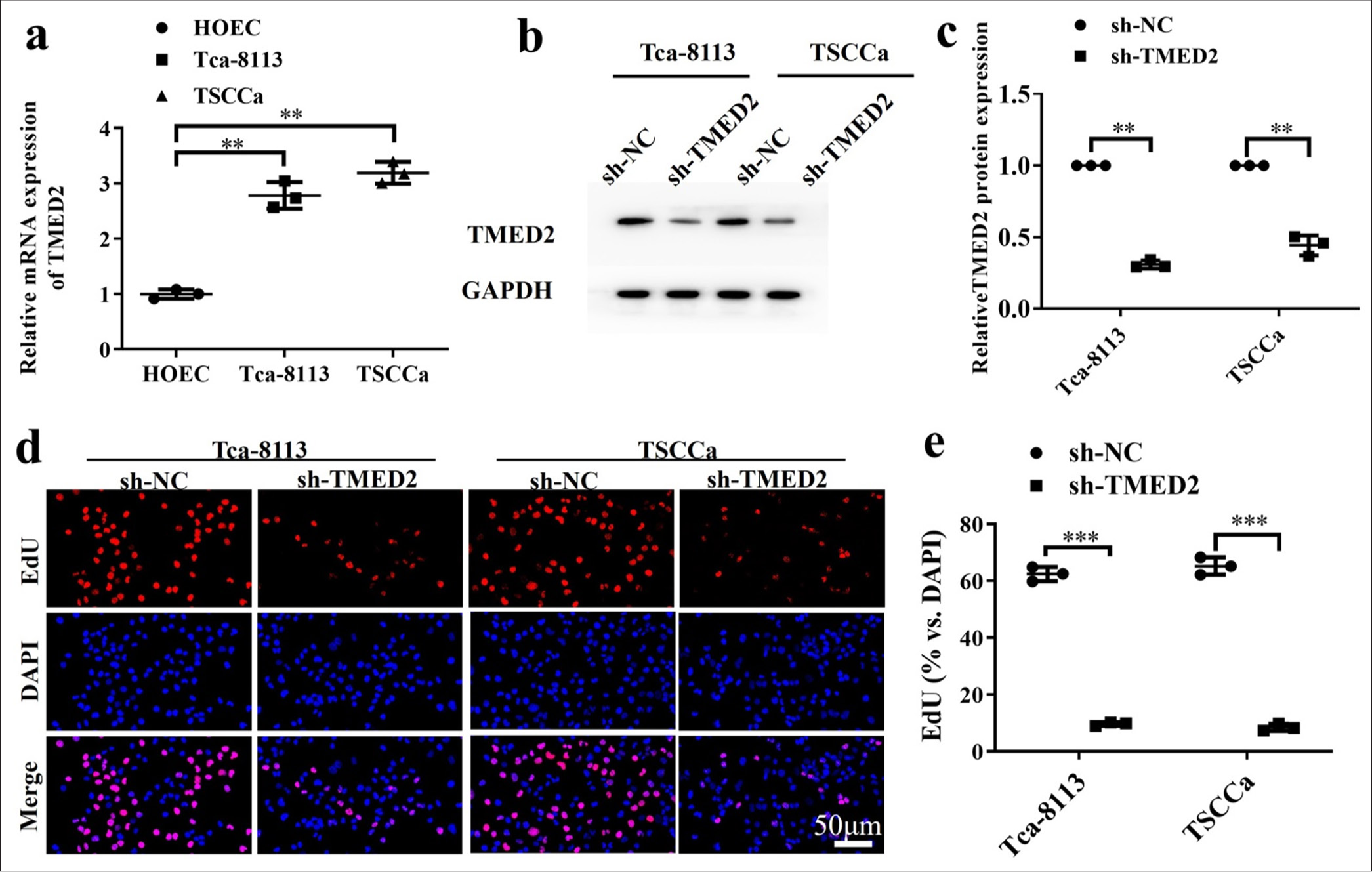
- TMED2 promotes proliferation of OC cells in vitro. (a) TMED2 expression levels in HOECs, Tca-8113, and TSCCa by qRTPCR. (b) TMED2 in OC cells. (c) TMED2 protein levels in OC cells. (d and e) Assessment of proliferation activity by EdU assay. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. TMED2: Transmembrane Emp24 domain containing 2, OC: Oral cancer, HOEC: Human oral epithelial cell, qRTPCR: Quantitative real-time polymerase chain reaction, sh-NC: negative control to TMED2 shRNA, sh-TMED2: TMED2 shRNA , GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, DAPI: 4’,6-diamidino-2-phenylindole, EdU: 5-ethynyl-2'-deoxyuridine.
TMED2 promotes migration and invasion of OC cells in vitro
The migration and invasion abilities of OC cells with sh-NC and sh-TMED2 were evaluated using Transwell assays. The cells in the sh-NC group were more capable of migrating and invading than those in the sh-TMED2 group [P < 0.01, Figure 2a-d].
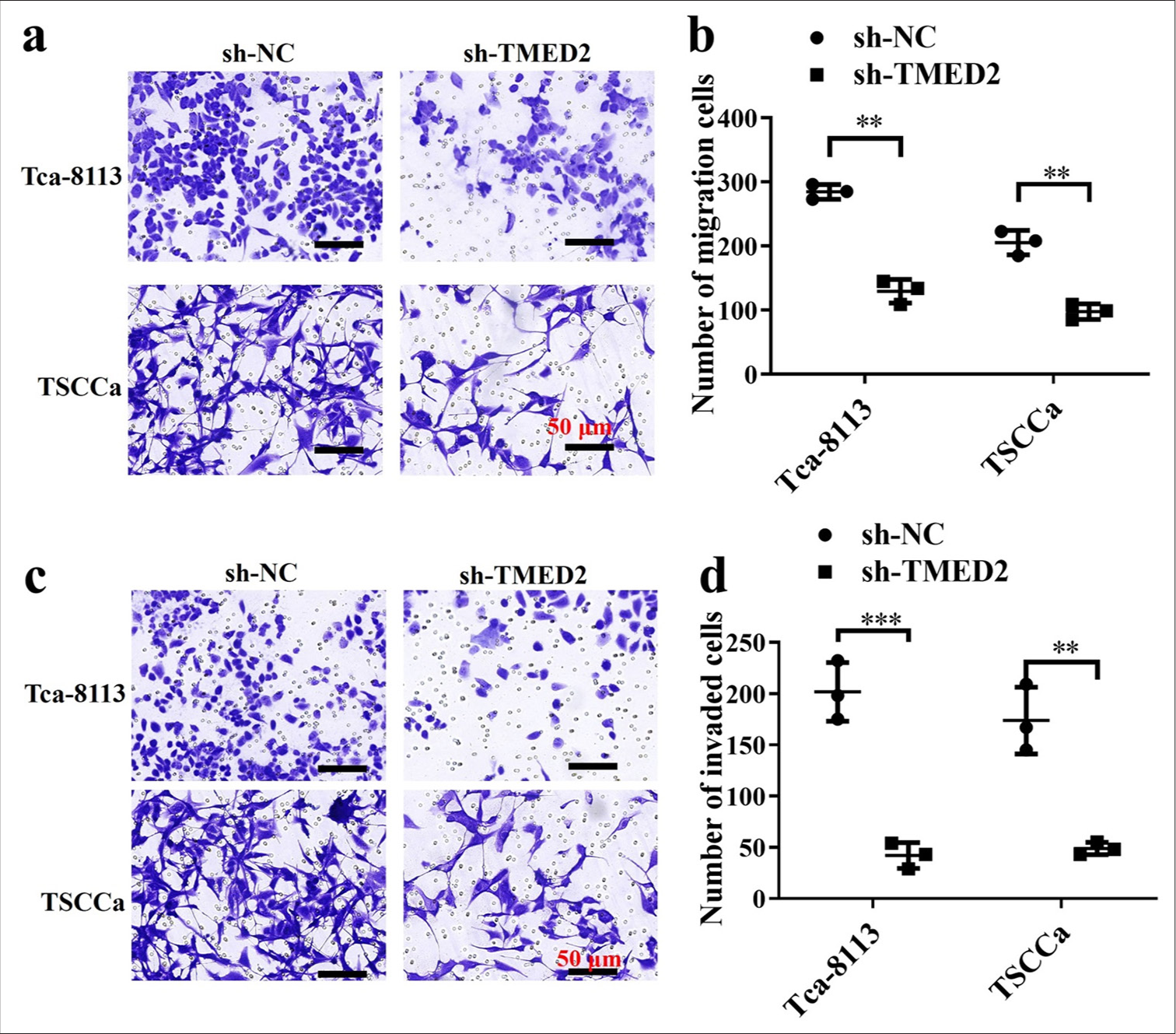
- TMED2 promotes migration and invasion of OC cells in vitro. (a and b) Migration of Tca-8113 and TSCCa cells transfected with sh-TMED2. (c and d) Invasion of Tca-8113 and TSCCa cells transfected with sh-TMED2. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001.
TMED2 promotes CSC-like characteristics and tumorigenicity in OC
SOX2, keratin 18 (KRT18), CD24 , and Nanog are widely used for identifying stem cell-like characteristics. Compared with those in the sh-NC group, SOX2, KRT18, CD24, and Nanog in the sh-TMED2 group were lower [P < 0.01, Figure 3a-d]. Therefore, the knockdown of TMED2 can reduce the stem cell-like characteristics of OC cells. The findings of the clonogenic experiment demonstrated that the cells in the shNC group produced more and larger colonies than the cells in the sh-TMED2 group (P < 0.01, Figure 3e and f]. Thus, silencing TMED2 expression can inhibit the clonogenic ability of OC cells.
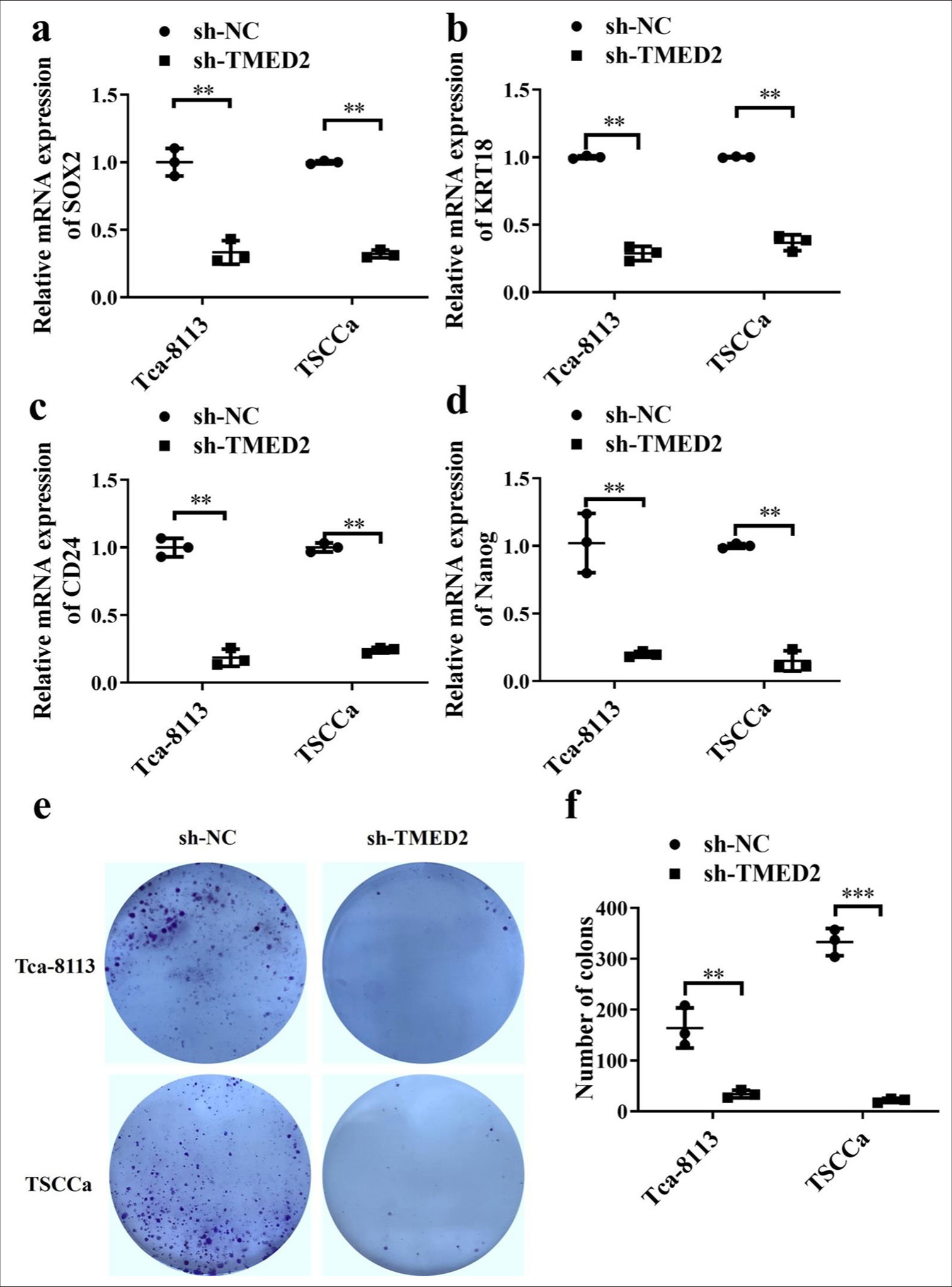
- TMED2 promotes CSC-like characteristics in OC. (a-d) Changes in SOX2, KRT18, CD24, and Nanog in OC cells. (e and f) Changes in clonogenic capacity in OC cells. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. CSC: Cancer stem cell, SOX2: SRY-box transcription factor 2, KRT18: Keratin 18.
TMED2 directly regulates the expression of GALNT7 in OC
The levels of GALNT7 in OC cells from the sh-NC and shTMED2 groups were measured by qRT-PCR. Compared with the sh-TMED2 group, the sh-NC group had a greater level of GALNT7, which suggested that silencing TMED2 downregulates GALNT7 expression [P < 0.01, Figure 4a]. The levels of GALNT7 protein in different treatment groups were also measured by Western blot. In OC cells, GALNT7 expression was considerably lower in the sh-TEMD2+OE-vector and sh-NC+OE-vector groups than in the shTMED2+OE-GALNT7 group (P < 0.05). Compared with that in the sh-TMED2+OE-vector group, the expression of GALNT7 in the sh-NC+OE-vector group was considerably higher [P < 0.01, Figure 4b and c]. CCK-8 results showed that the cell proliferation activity of the sh-NC+OE-GALNT7 group was significantly higher than that of the sh-NC+OE-vector and sh-TEMD2+OE-vector groups (P < 0.05). The sh-NC+OE-vector group was significantly higher than the sh-TEMD2+OE-vector group in terms of cell proliferation activity [P < 0.01, Figure 4d]. Figure 4e-h shows the results of Transwell experiment. Compared with those in the shTEMD2+OE-vector and sh-NC+OE-vector groups, the OC cells treated with GALNT7 overexpression exhibited considerably increased migration and invasion capabilities (P < 0.01). The sh-NC+OE-vector group had significantly higher capabilities than the sh-TEMD2+OE-vector group (P < 0.001).
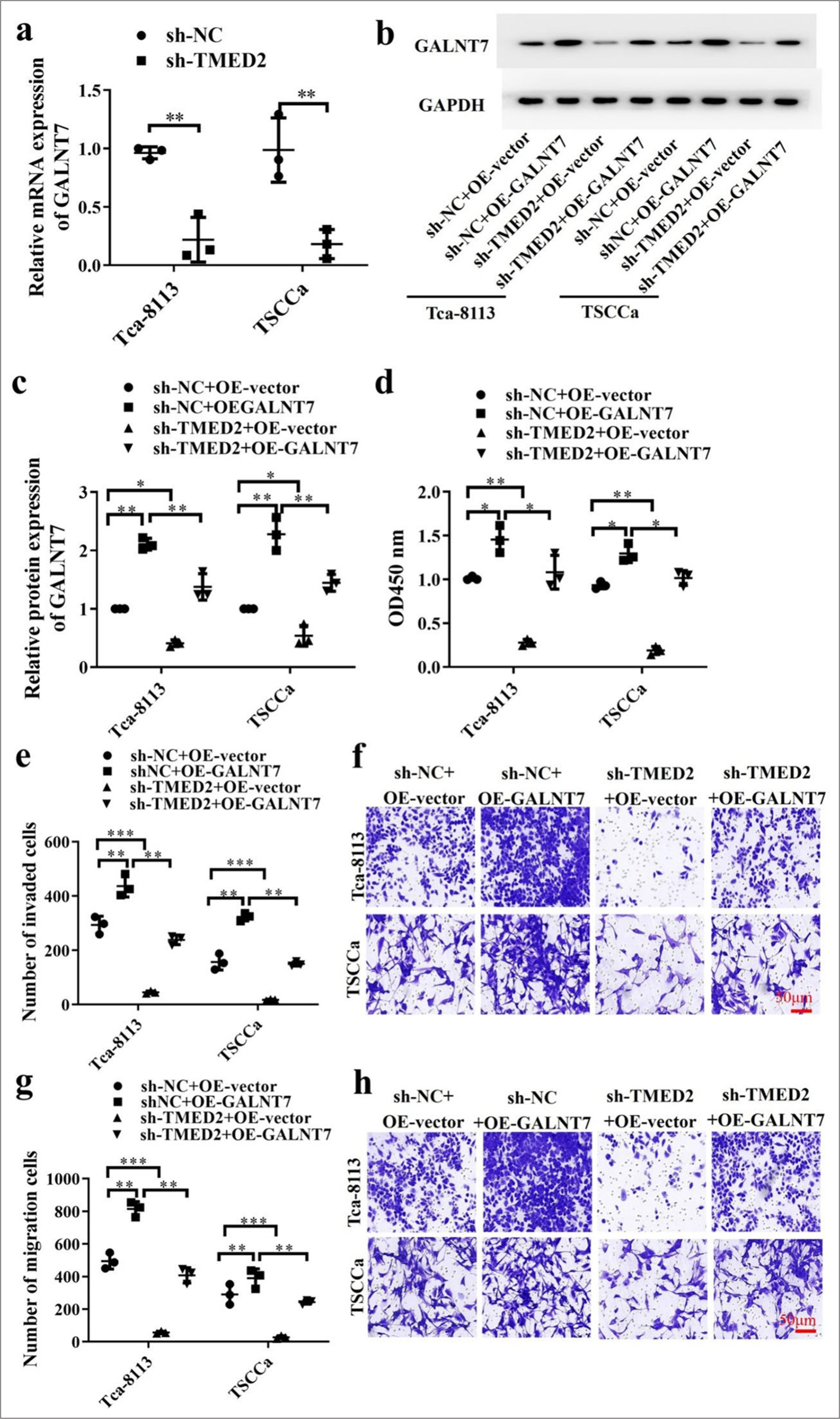
- TMED2 regulates GALNT7 expression in OC. (a) Measurement of GALNT7 in OC cells with sh-TMED2. (b) Measurement of GALNT7 protein levels. (c) Statistical analysis of GALNT7 protein levels. (d) Assessment of proliferation activity by CCK-8 assay. (e-h) Migration and invasion of OC cells transfected with sh-NC+OE-vector, sh-NC+OE-GALNT7, sh-TMED2+OE-vector, or sh-TMED2+OE-GALNT7. Scale bar: 50 μm. n = 3, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. GALNT7: Polypeptide-N-acetylgalactosaminyltransferase 7, CCK-8: Cell counting kit-8.
TMED2 regulates GALNT7 to participate in CSC-like characteristics
In OC cells, stem cell markers SOX2, OCT4, CD24, Nanog, myelocytomatosis oncogene (MYC), and B-cell-specific Moloney murine leukemia virus insertion site 1 (BMI1) in the sh-NC+ OE-GALNT7 group were increased compared with those in the sh-NC+OE-vector and sh-TMED2+OE-vector groups (P < 0.05). In addition, the sh-TEMD2+OE-vector group had substantially lower stem cell markers than the sh-NC+OE-vector group [P < 0.01, Figure 5a-f]. This result indicates that the regulation of TMED2 and GALNT7 plays an important role in maintaining stem cell-like characteristics. The results of the clonogenic assay also revealed that, in contrast to those in the sh-NC+OE-GALNT7 group, fewer colonies were produced in the sh-TMED2+OE-vector and sh-NC+OE-vector groups (P < 0.01). Moreover, the sh-TMED2+OE-vector group had a considerably smaller number of colonies than the sh-NC+OE-vector group [P < 0.001, Figure 5g and h]. Information from the TCGA database indicated that TMED2 and GLANT7 were positively correlated with poor prognosis [Figure 5i and j].
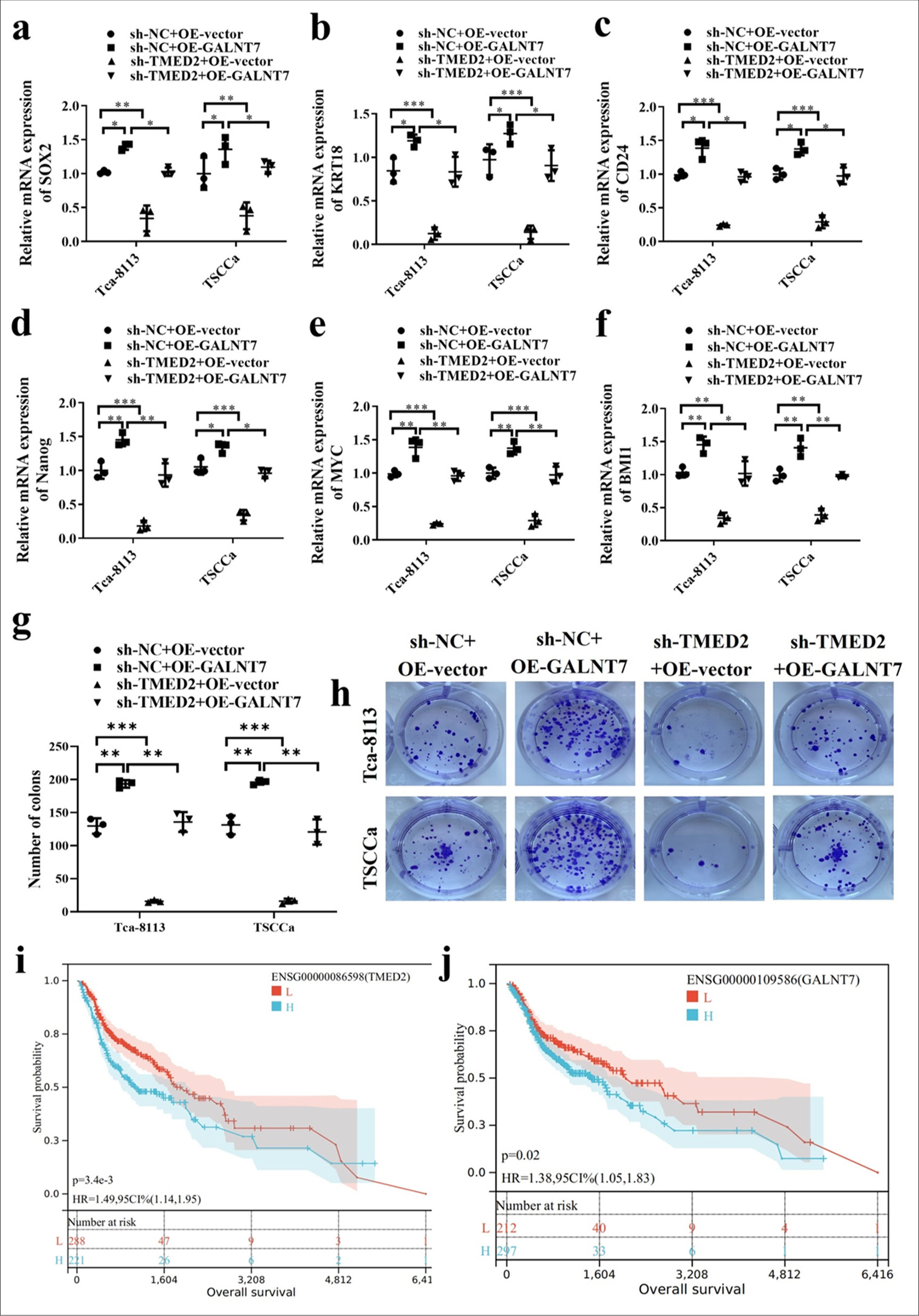
- TMED2 regulates CSC-like characteristics through GALNT7. (a-f) Changes in stem cell markers SOX2, OCT4, CD24, Nanog, MYC, and BMI1 in OC cells. (g and h) Soft agar colony formation assay in OC cells. (i and j) Survival curves of TMED2 and GALNT7. n = 3, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. MYC: myelocytomatosis oncogene, BMI1: B-cell-specific Moloney murine leukemia virus insertion site 1.
TMED2 regulates GALNT7 to inhibit tumor growth and metastasis in xenograft tumor models
We introduced Tca-8113 cells into the right axillary fossa of the mouse to examine the function of TMED2 in impeding tumor growth and metastasis in vivo. The findings demonstrated a significant decrease in tumor volume and weight in the Control+sh-NC+OE-vector and Control+shTMED2+OE-GLANT7 groups as compared to the Control+sh-NC+OE-GALNT7 group [P < 0.01, Figure 6a-c]. H E staining showed that the Control+sh-NC+OE-GALNT7 group had larger and diffusely distributed tumor cells, while those in the Control+sh-NC+OE-vector and Control+shTMED2+OE-GLANT7 groups were smaller with enlarged cell spaces [Figure 6d]. In addition, pulmonary metastasis was significantly reduced after silencing TMED2 expression [P < 0.001, Figure 6e and f]. Ki67 immunohistochemical results showed that the Ki67 level of the Control+shNC+OE-GALNT7 group was significantly increased. Silencing GALNT7 expression reversed this trend [P < 0.001, Figure 6g and h]. Finally, the mRNA levels of CD24, MYC, and BMI1 were determined by qRT-PCR [P < 0.01, Figure 6i-k]. Western blot was conducted to evaluate the levels of SOX2, OCT4, and Nanog protein expression. Their expression levels were significantly increased in the Control+sh-NC+OE-GALNT7 group but were significantly decreased in the Control + sh-TMED2+OE-GLANT7 group [P < 0.05, Figure 6l and m]. These results suggest that TEMD2 inhibits tumor growth and metastasis in vivo by regulating GALNT7. The results of previous in vitro experiments further demonstrated the link between TMED2 and GALNT7 and their role in OC.
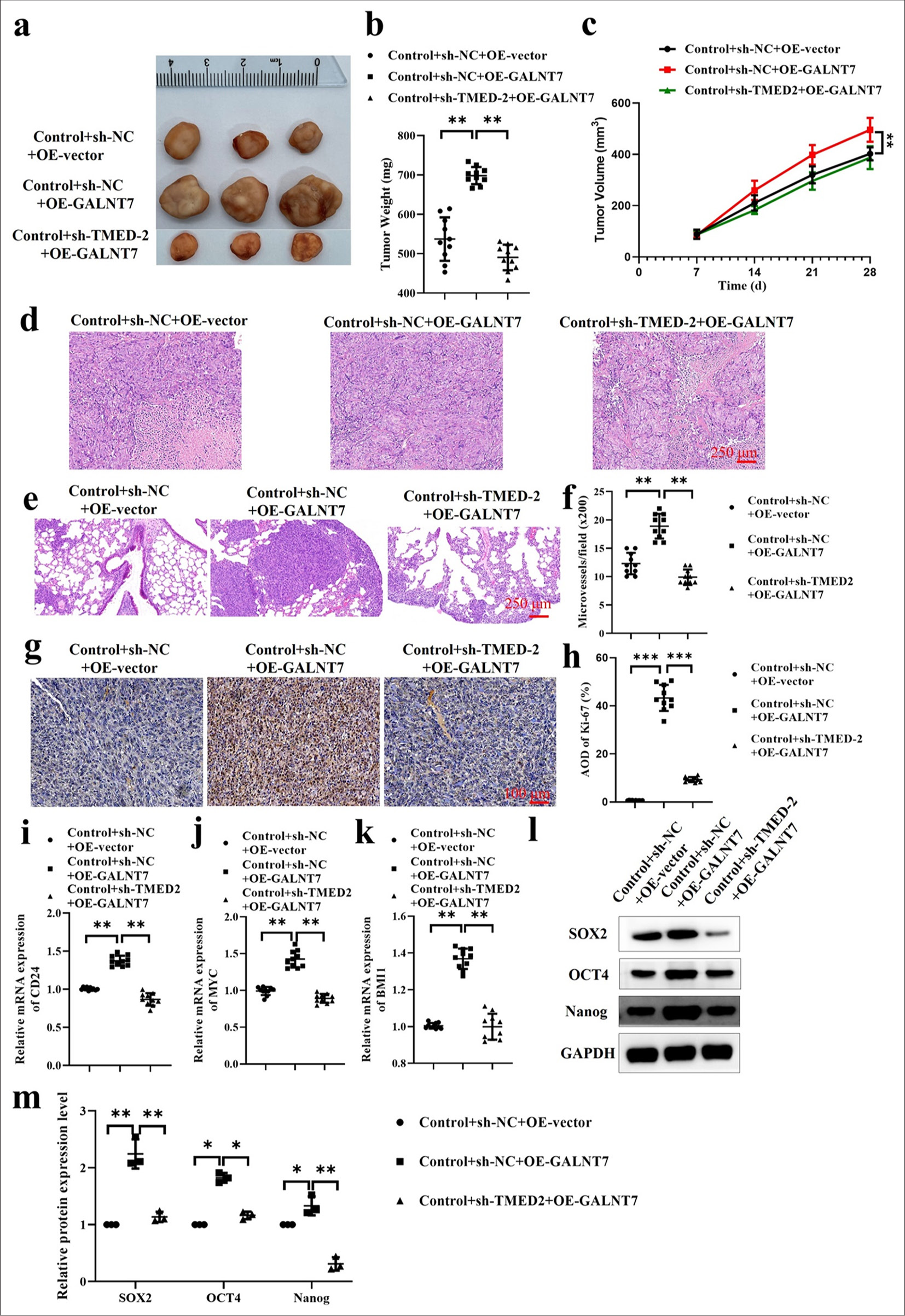
- TMED2 regulates GALNT7 to inhibit tumor growth and metastasis in xenograft tumor models. (a) Tumor xenograft. (b) Tumor weight. (c) Tumor volume. (d) H and E staining of tumor. (e and f) H E staining of lung. (g and h) Ki67 immunohistochemical staining of tumor. (i-k) The mRNA expression levels of CD24, MYC, and BMI1. (l and m) Protein expression levels of SOX2, OCT4, and Nanog. n = 10, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. H and E: Hematoxylin and eosin.
DISCUSSION
OC involves multiple complex molecular mechanisms in its development and progression.[19] Their capacity for self-renewal and multidirectional differentiation promotes tumor growth, recurrence, and metastasis. Thus, CSCs are regarded as essential components in OC.[7,20] Consequently, research into the traits and control systems of CSCs is crucial to managing OC.[9,21] This study demonstrates that TMED2 plays a critical role in OC by promoting the transcription of GALNT7, which enhances the stem-like properties and tumorigenicity.
TMED2 is a transmembrane protein involved in the transport process between the endoplasmic reticulum and Golgi apparatus, and it plays an important role in various cancers.[22] It is strongly linked to the development of tumors and their malignant transformation, and it is substantially expressed in OC. Proliferation, migration, and invasion are key features of cancer development and metastasis, and the upregulation of TMED2 appears to be associated with the enhancement of these features. These findings are consistent with those in previous studies, which indicate the role of TMED2 in promoting cell proliferation in various cancers. CSCs are considered the driving force behind tumor initiation and progression.[23] The upregulation of TMED2 is associated with stem cell markers and enhanced tumorigenicity. Therefore, TMED2 may enhance the tumorigenicity of OC by promoting stem-like properties. This finding provides new targets and strategies for the treatment of OC. Silencing TMED2 can reduce the occurrence of OC, which further confirms its important role in OC and provides support for TMED2 as a potential therapeutic target. Interfering with TMED2 may effectively inhibit the invasion abilities of cancer cells and reduce the stem-like properties and tumorigenicity, which effectively suppress the development and metastasis of OC.
GALNT7 is a glycosyltransferase that has been shown to play a crucial role in various cancers.[17,24] In addition, studies have found that TMED2 and GALNT7 are important markers,[25] and our results also prove this point. The upregulation of TMED2 increases the transcription level of GALNT7. Further works have revealed that the upregulation of GALNT7 is closely associated with enhanced stem-like properties and tumorigenicity in OC. Improved stem-like properties include increased self-renewal, proliferation, invasion, and drug resistance capabilities. These findings suggest that TMED2 enhances the stem-like properties of OC by promoting the transcription of GALNT7, which drives tumor initiation and progression. This study not only proves that TMED2 and GALNT7 are important markers but also further discusses the relationship between them.
TMED2 and GALNT7 play important roles in OC, which makes them potential targets for developing novel therapeutic approaches. One potential application is to inhibit the properties and tumorigenicity of CSCs by targeting TMED2 or GALNT7. Previous research has demonstrated that tumor growth and metastasis can be inhibited by blocking TMED2 or GALNT7. Therefore, the development of inhibitors targeting TMED2 and GALNT7 may become a novel strategy for the treatment of OC. In addition, the expression of TMED2 and GALNT7 has been found to be associated with the prognosis of OC. Therefore, TMED2 and GALNT7 can serve as potential biomarkers for prognosis assessment and treatment response monitoring in OC.
In this study, the previous relationship between TEMD2 and GALNT7 and their role in OC were discussed only in cells and mice. In future works, we will use a wider variety of sample types beyond clinical patient samples to conduct in-depth exploration.
SUMMARY
TMED2 enhances stem-like properties and tumorigenicity in OC by promoting the transcription of GALNT7. It also inhibits tumor growth and metastasis in vivo. This discovery offers new insights into the processes underlying the onset and advancement of OC and identifies possible targets for innovative treatment approaches. Further research will elucidate the specific interaction mechanisms between TMED2 and GALNT7 and evaluate their potential clinical applications.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study were available from the corresponding author on reasonable request.
ACKNOWLEDGMENT
Not applicable.
ABBREVIATIONS
AKT: Protein kinase B
BMI1: B-cell-specific Moloney murine leukemia virus insertion site 1
CCK-8: Cell counting kit-8
CSCs: Cancer stem cells
DMEM: Dulbecco’s modified eagle medium
EdU: 5-ethynyl-2’-deoxyuridine
GALNT7: Polypeptide-N-acetylgalactosaminyltransferase 7
H and E: Hematoxylin and eosin
KRT18: Keratin 18
OC: Oral cancer
SOX2: SRY-box transcription factor 2
PI3K: Phosphoinositide 3-kinase
STR: Short tandem repeat
TMED2: Transmembrane Emp24 domain containing 2.
AUTHOR CONTRIBUTIONS
YHW and SCZ: Designed the study; all authors conducted the study; GMZ: Collected and analyzed the data; GMZ, YHW and SCZ: Participated in drafting the manuscript, and all authors contributed to critical revision of the manuscript for important intellectual content. All authors gave final approval of the version to be published. All authors participated fully in the work, take public responsibility for appropriate portions of the content, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or completeness of any part of the work are appropriately investigated and resolved.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study has been approved by the ethics committee of Nanjing Integrated Traditional Chinese and Western Medicine Hospital, Affiliated Hospital of Nanjing University of Chinese Medicine, approval No. 2024050. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Nanjing Integrated Traditional Chinese and Western Medicine Hospital, Affiliated Hospital of Nanjing University of Chinese Medicine. This article does not involve patients, so informed consent is not required.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- Oral cancer and precancer: A narrative review on the relevance of early diagnosis. Int J Environ Res Public Health. 2020;17:9160.
- [CrossRef] [PubMed] [Google Scholar]
- Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127-35.
- [CrossRef] [PubMed] [Google Scholar]
- Microbiota and oral cancer as a complex and dynamic microenvironment: A narrative review from etiology to prognosis. Int J Mol Sci. 2022;23:8323.
- [CrossRef] [PubMed] [Google Scholar]
- The matrix revolution: Matricellular proteins and restructuring of the cancer microenvironment. Cancer Res. 2020;80:2705-17.
- [CrossRef] [PubMed] [Google Scholar]
- Cancer microenvironment defines tumor-infiltrating lymphocyte density and tertiary lymphoid structure formation in laryngeal cancer. Head Neck Pathol. 2023;17:422-32.
- [CrossRef] [PubMed] [Google Scholar]
- The current markers of cancer stem cell in oral cancers. Life Sci. 2020;249:117483.
- [CrossRef] [PubMed] [Google Scholar]
- Cancer stem cell self-renewal as a therapeutic target in human oral cancer. Oncogene. 2019;38:5440-56.
- [CrossRef] [PubMed] [Google Scholar]
- Oral cancer: Genetics and the role of precision medicine. Surg Oncol Clin. 2020;29:127-44.
- [CrossRef] [PubMed] [Google Scholar]
- The stress hormone norepinephrine promotes tumor progression through β2-adrenoreceptors in oral cancer. Arch Oral Biol. 2020;113:104712.
- [CrossRef] [PubMed] [Google Scholar]
- TMED2-ALK, a novel ALK fusion gene identified in a patient with lung adenocarcinoma. J Thorac Oncol. 2020;15:e37-9.
- [CrossRef] [PubMed] [Google Scholar]
- The circular RNA CDR1as regulate cell proliferation via TMED2 and TMED10. BMC Cancer. 2020;20:312.
- [CrossRef] [PubMed] [Google Scholar]
- TMED2 binding restricts SMO to the ER and Golgi compartments. PLoS Biol. 2022;20:e3001596.
- [CrossRef] [PubMed] [Google Scholar]
- Transmembrane p24 trafficking protein 2 regulates inflammation through the TLR4/NF-κB signaling pathway in lung adenocarcinoma. World J Surg Oncol. 2022;20:32.
- [CrossRef] [PubMed] [Google Scholar]
- Expression and importance of TMED2 in multiple myeloma cells. Cancer Manag Res. 2020;12:12895-903.
- [CrossRef] [PubMed] [Google Scholar]
- TMED2 induces cisplatin resistance in breast cancer via targeting the KEAP1-Nrf2 pathway. Curr Med Sci. 2023;43:1023-32.
- [CrossRef] [PubMed] [Google Scholar]
- The role of GALNT7 as a potential diagnostic marker in prostate cancer. Nat Rev Urol. 2023;20:198.
- [CrossRef] [Google Scholar]
- Upregulation of GALNT7 in prostate cancer modifies O-glycosylation and promotes tumour growth. Oncogene. 2023;42:926-37.
- [CrossRef] [PubMed] [Google Scholar]
- SPDEF enhances cancer stem cell-like properties and tumorigenesis through directly promoting GALNT7 transcription in luminal breast cancer. Cell Death Dis. 2023;14:569.
- [CrossRef] [PubMed] [Google Scholar]
- Photodynamic therapy in oral cancer: A review of clinical studies. Med Oncol. 2023;40:91.
- [CrossRef] [PubMed] [Google Scholar]
- Effects of 5-ALA mediated photodynamic therapy in oral cancer stem cells. J Photochem Photobiol B. 2022;235:112552.
- [CrossRef] [PubMed] [Google Scholar]
- EGFR enhances the stemness and progression of oral cancer through inhibiting autophagic degradation of SOX2. Cancer Med. 2020;9:1131-40.
- [CrossRef] [PubMed] [Google Scholar]
- TMED family genes and their roles in human diseases. Int J Med Sci. 2023;20:1732-43.
- [CrossRef] [PubMed] [Google Scholar]
- Cancer stem cells as key drivers of tumour progression. J Biomed Sci. 2018;25:20.
- [CrossRef] [PubMed] [Google Scholar]
- Knockdown of GALNT7 promotes cell apoptosis and autophagy of breast cancer cells by inactivation of STAT3. Eur J Gynaecol Oncol. 2022;43:79-85.
- [Google Scholar]
- Biomarkers in otorhinolaryngology In: Biomarkers in medicine. Sharjah, UAE: Bentham Science; 2022. p. :276-308.
- [CrossRef] [Google Scholar]








