Translate this page into:
Scavenger receptor class B member 1 promotes lung cancer growth and metastasis through enhanced twist family BHLH transcription factor 1 signaling in vitro and in vivo: Exploration of RPPNs as a therapeutic Strategy

*Corresponding author: Hong Bian, Department of Thoracic Surgery, Southern University of Science and Technology Hospital, Guangdong, China. szbianhong@outlook.com
-
Received: ,
Accepted: ,
How to cite this article: Liu X, Bian H, Shi Y, Du T. Scavenger receptor class B member 1 promotes lung cancer growth and metastasis through enhanced twist family BHLH transcription factor 1 signaling in vitro and in vivo: Exploration of RPPNs as a therapeutic Strategy. CytoJournal. 2025;22:23. doi: 10.25259/Cytojournal_222_2024
Abstract
Objective
Lung cancer remains a leading cause of cancer-related mortality worldwide. Although scavenger receptor class B member 1 (SCARB1), a crucial cell surface receptor, plays a vital role in various cancers, its function in lung cancer remains incompletely elucidated. This study aims to investigate the role and molecular mechanisms of SCARB1 in lung cancer progression and develop a novel SCARB1-targeted nanoparticle drug delivery system.
Material and Methods
We analyzed SCARB1 expression levels in lung cancer tissues and their correlation with patient prognosis using the Cancer Genome Atlas database. In vitro experiments, including quantitative real-time polymerase chain reaction, Western blot, 5-ethynyl-2'-deoxyuridine, colony formation, and Transwell analyses, were conducted to study the effects of SCARB1 on lung cancer cell proliferation, invasion, and migration. A lung metastasis model was established through tail vein injection to evaluate the role of SCARB1 in promoting lung cancer metastasis in vivo. We also developed red blood cell membrane-coated poly (lactic-co-glycolic acid) nanocarriers loaded with paclitaxel (RPPNs) and assessed their effect on SCARB1 expression and lung cancer progression.
Results
SCARB1 was overexpressed in human lung cancer tissues and significantly associated with poor patient prognosis. In vitro experiments confirmed that silencing SCARB1 inhibited lung cancer cell growth, invasion, and migration. SCARB1 overexpression promoted lung cancer cell proliferation, migration, and epithelial–mesenchymal transition through twist family BHLH transcription factor 1 (Twist1) activation. In vivo experiments further validated the crucial role of SCARB1 in promoting lung cancer metastasis. The developed RPPNs effectively suppressed SCARB1 expression in lung cancer and demonstrated superior inhibitory effects compared with traditional RPPNs. However, SCARB1 overexpression partially antagonized the antimetastatic effects of RPPNs.
Conclusion
This work elucidates, for the 1st time, the molecular mechanism by which SCARB1 promotes lung cancer growth and metastasis through the activation of the Twist1 signaling pathway and develops a novel SCARB1-targeted nanoparticle drug delivery system, namely RPPNs. The findings of this work not only deepen our understanding of the molecular mechanisms underlying lung cancer progression but also provide new strategies for lung cancer diagnosis and treatment.
Keywords
Lung cancer
Nanoparticle drug delivery
Scavenger receptor class B member 1
Twist family bHLH transcription factor 1
INTRODUCTION
Lung cancer is one of the malignant neoplasms with the highest incidence and mortality rates globally and poses a severe threat to human health.[1,2] Although its diagnosis and treatment have advanced considerably in recent years, its 5-year survival rate remains low because of tumor invasion, metastasis, and therapeutic resistance.[3,4] Investigating the molecular mechanisms of lung cancer development and progression and exploring new therapeutic targets and strategies are crucial to enhance the survival rates and quality of life of patients with lung cancer.[5,6]
Scavenger receptor class B member 1 (SCARB1) is a multifunctional transmembrane glycoprotein that was first recognized as a high-density lipoprotein receptor involved in reverse cholesterol transport.[7,8] It is abnormally expressed in multiple tumors and remarkably linked to tumor progression and prognosis.[9,10] In breast cancer, high SCARB1 expression is correlated with tumor invasiveness and poor prognosis.[11] In addition, SCARB1 promotes the metastasis of various tumor cells, such as nasopharyngeal carcinoma[10] and human osteosarcoma cells.[12] The role and molecular mechanisms of SCARB1 in lung cancer are not yet fully understood. Twist family bHLH transcription factor 1 (Twist1) is a crucial transcription factor that plays a pivotal role in embryonic development and tissue remodeling. It is a key regulator of epithelial-mesenchymal transition (EMT) in tumors and facilitates tumor cell invasion and metastasis.[13,14] Multiple studies have shown that Twist1 is overexpressed in lung cancer and associated with tumor progression and poor prognosis.[15,16] In addition, Twist1 promotes the development and metastasis of breast cancer tumors. However, the upstream regulatory mechanisms of Twist1 in lung cancer and its association with SCARB1 warrant further exploration.
Nanocarrier drug delivery systems have demonstrated immense potential in tumor therapy.[17,18] Poly (lactic-coglycolic acid) (PLGA) nanoparticles have gained attention for their outstanding biocompatibility and biodegradability.[19] Researchers have developed various biomembrane-coated nanocarrier systems to further enhance the targeting ability and biocompatibility of nanocarriers.[20] Red blood cell membrane (RBCM)-coated nanocarriers have gained particular favor because of their unique advantages.[21,22] Paclitaxel (PTX) is an essential broad-spectrum antitumor medication used in lung cancer therapy.[23] However, its clinical application is limited by factors, such as poor water solubility and low bioavailability. Loading PTX into nanocarriers can effectively address these issues and improve its therapeutic efficacy.[24] Our previous studies revealed that PTX can inhibit SCARB1 expression and provide new insights for developing targeted therapeutic strategies against SCARB1.
This study explores the role and molecular mechanisms of SCARB1 in lung cancer progression and aims to develop a novel SCARB1-targeted nanoparticle drug delivery system. We propose that SCARB1 could facilitate lung cancer progression and metastasis through the Twist1 signaling pathway and that RBCM-coated PLGA nanocarriers loaded with PTX RPPNs might enhance antitumor efficacy by targeting SCARB1.
This study clarifies the role and molecular mechanisms of SCARB1 in lung cancer progression to provide a theoretical basis for identifying new diagnostic biomarkers and therapeutic targets. It also develops a SCARB1-targeted nanoparticle drug delivery system and offers innovative strategies to enhance lung cancer treatment efficacy. Its results are expected to enhance early diagnosis, prognosis assessment, and personalized treatment for lung cancer, thereby improving patient survival rates and quality of life.
MATERIAL AND METHODS
Bioinformatic analysis
The TNMplot database (https://tnmplot.com/analysis/) was utilized to analyze the correlation between the expression of SCARB1 and clinicopathological features of lung cancer pathological stages. The Sangerbox3. 0 database (http://sangerbox.com/) and data from the Cancer Genome Atlas (TCGA) were used to examine the association between SCARB1 expression and prognosis in patients with lung cancer. The correlation between SCARB1 expression levels and tumor stemness was then analyzed. If the data fit the normal distribution, the t-test was used to compare the difference between the two groups. If the data conformed to a non-normal distribution, a rank-sum test was used to compare the difference between the two groups. Bilateral P < 0.05 was considered statistically significant.
Cell culture
A549 cells (BFN60800665) were obtained from American Type Culture Collection (Manzas, Virginia, the USA) and cultured in complete Dulbecco’s Modified Eagle Medium (31600, Solarbio, Beijing, China) with 10% fetal bovine serum (FBS, S9020, Solarbio, Beijing, China) at 37°C and 5% Carbon dioxide (CO2). The cells were mycoplasma-free, and short tandem repeat analysis revealed that they were derived from their parental cells. A549 cells in the logarithmic growth phase were plated into six-well plates at a density of 4 × 105 cells per well. Transfection was conducted at 70% cell confluence by following the instructions included with the HighGeneplus transfection reagent (RM09014, ABclonal, Hubei, China). Lung cancer cells were divided into the SCARB1#1 siRNA (si-SCARB1#1), SCARB1#2 siRNA (si-SCARB1#2), overexpression (OE)- SCARB1, and negative control to SCARB1#1 or SCARB1#2 siRNA (si-NC) groups For nanomaterial evaluation, the groups were divided into the control, PLGA-PTX-NPs (PPNs) treatment, and RPPNs treatment groups. PPNs (10 μg/mL) or RPPNs (10 μg/mL) were added for 24 h.
SCARB1#1 siRNA: sense: 5'-CAAGUUCGGAUUAUUU GCUTT-3', antisense: 5'-AGCAAAUAAUCCGAACUU GTT-3'
SCARB1#2 siRNA: sense: 5'-GGACAGAUAUC AUCAA CGA-3, antisense: 5'-UCGUUGAUG AUAUCU GU-3'
SCARB1 OE plasmid: sense: 5'-ATTGAATTCTGA AAACAAGTAATAGCTTTGGCTG-3', antisense: 5'-GCGGCCGCGCATTTGTTAATGGTATTTA-3'.
Construction of the SCARB1 OE cell line
A549 lung adenocarcinoma cells were revived and passaged. A transfection reagent and Opti- Minimal Essential Medium were added to transfect A549 cells with the OE plasmid. The cells were incubated at 37°C with 5% CO2 for 8 h before medium replacement. Transfection efficiency was assessed after 24 h. After successful transfection was verified, the medium was replaced, and the cells were incubated for another 24 h.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of relative gene expression
Total ribonucleic acid (RNA) was extracted using TRIeasy™ Total RNA Extraction Reagent (10606ES60, Yeasen, Shanghai, China). A total of 1 μg of RNA was extracted in accordance with the instructions of the RNA rapid extraction kit (AC0202, SparkJade, Shandong, China). qRT-PCR analysis was conducted in accordance with the protocol of the All-in-One™ First Strand complementary desoxyribonucleic acid (cDNA) Synthesis Kit (QP007, iGeneBio, Guangdong, China). The primer sequences are shown in Table 1. The polymerase chain reaction (PCR) conditions included 95°C for 30 s; 95°C for 5 s, 63°C for 20 s, and 72°C for 20 s for 40 cycles. Relative quantitative research software (7300 Sequence Detection system, version 1.4, Applied Biosystems) was used to perform relative quantification through the comparative Ct method (2−ΔΔCt method). The sequence in Table 1 was normalized to the amount of endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and relative to that of the calibrator.
| Primer | Primer sequence (5'–3') |
|---|---|
| SCARB1 | CCTATCCCCTTCTATCTCTCCG |
| GGATGTTGGGCATGACGATGT | |
| Twist1 | GTCCGCAGTCTTACGAGGAG |
| GCTTGAGGGTCTGAATCTTGCT | |
| E-cadherin | CCAGACCCAACGCGCGACCCGGACCTCCC |
| TTTTTTTTTTTTTTTTTT TTTGGCCAGAAAGCAA | |
| Vimentin | GCGTGACGTACGTCAGCAATATGA |
| GTTCCAGGGACTCATTGGTTCCTT | |
| GAPDH | GACAACTTTGGCATCGT |
| ATGCAGGGATTGATGTTCTGG |
SCARB1: Scavenger receptor class B member 1, Twist1: Twist family bHLH transcription factor 1, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Western blot analysis of SCARB1 and Twist1 protein expression levels
The culture dish was aspirated to remove the cell culture medium and added with 100 μL of cell lysis buffer (R0010, Solarbio, Beijing, China). The mixture was chilled on ice for 20 min. The supernatant was collected after centrifugation at 12 000 rpm for 10 min, and protein concentration was determined. Proteins were denatured, loaded, and separated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After membrane transfer, the blot was blocked with 3% skimmed milk for 1 h, then incubated overnight at 4°C with primary antibodies against SCARB1 (#90332), Twist1 (#90445), and GAPDH (#2118), each at a 1:1000 dilution. The membrane was washed and then incubated with secondary antibodies (1:2000 dilution, #7074) at 37°C for 1.5 h. All antibodies were purchased from CST (Ma, BSN, the USA). After being washed again, the blot was added with enhanced chemiluminescence (BL520b, Biosharp Life Science, Hefei, Anhui, China) chemiluminescent substrate and imaged using a gel imaging system (Image Quant LAS4000, GE Healthcare, Chicago, IL, the USA). Band intensities were analyzed using Quantity One software (4.6.2, Bio-RAD, Hercules, CA, the USA).
5-Ethynyl-2'-deoxyuridine (EdU) cell proliferation assay
A549 cells were seeded into 24-well plates at a density of 1 × 105 cells per well and cultured for 24 h before being grouped and transfected. After 24 h, the cells were added to 10 μmol/L EdU solution (C0081S, Beyotime Biotechnology, Shanghai, China) and incubated for 2 h, then stained with 4',6-diamidino-2-phenylindole (C1005, Beyotime Biotechnology, Shanghai, China) for 10 min. The cells were observed and photographed under a fluorescence microscope (CX41-32RFL, Olympus, Tokyo, Japan) in accordance with the instructions of the EdU cell proliferation detection kit. The EdU-positive rate (%) was determined by dividing the number of EdU-positive cells by the total number of cells and multiplying by 100.
Colony formation assay
Cells from each group were trypsinized, thoroughly resuspended, and counted. The cells (1 × 103) were seeded into six-well plates, and the medium was refreshed every 3 days to monitor cell growth. The culture was terminated; after 2 weeks once, visible colonies had formed. The previous medium was removed, and the cells were washed twice with phosphate-buffered saline (PBS). The cells were treated with 4% paraformaldehyde and 0.5% crystal violet (G1065, Solarbio, Beijing, China) for 15 min each, then air dried. Colony numbers were counted using ImageJ software (v1.3.4, National Institutes of Health, Bethesda, MD, the USA) from photographs taken against a white background.
Transwell migration and invasion assays
Cells from each group were prepared, and a 200 μL cell suspension (3 × 104 cells/mL) was seeded into the upper chamber of a Matrigel-coated Transwell insert. The lower chamber was filled with 600 μL of a medium containing 30% FBS. Following 24 h of incubation at 37°C, cells that had penetrated the chamber were fixed using 4% paraformaldehyde and stained with 0.5% crystal violet for 20 min. The number of invaded cells was counted, and the cells were photographed under an optical microscope (DMi8, Zeiss, Oberkochen, Baden-Württemberg, Germany).
Development of a lung metastasis model through tail vein injection
Twenty-four female BALB/c nude mice, aged 4–6 weeks, were obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd. and maintained in a specific pathogen-free barrier environment at the Department of Laboratory Animal Science. The 3R principles and animal welfare guidelines were strictly followed. The Institutional Animal Care and Use Committee approved the use of mice in this study. A549 cells in the logarithmic growth phase were processed through digestion, centrifugation, and resuspension in PBS to prepare a 1 × 107/mL cell suspension. The cell suspension (100 μL) was administered to each nude mouse through the tail vein. At 6 weeks after injection, mice were sacrificed by neck dislocation, and their lungs were collected for hematoxylin and eosin (H&E) staining to examine lung metastases. RPPNs were intravenously administered at 10 mg/kg twice weekly for 6 weeks.
H&E staining of lung tissue
Lung tissue samples were fixed in 10% formalin for 7 days and subjected to routine paraffin embedding. Tumor tissue samples were deparaffinized, hydrated, stained with hematoxylin, differentiated and blued, stained with eosin, and dehydrated. The tissue specimens were sectioned using a paraffin microtome, air-dried, and mounted with neutral balsam. H&E staining (S0105S, Solarbio, Beijing, China) was used to observe pathological changes in lung tissue metastatic foci.
Immunohistochemical (IHC) staining
The EnVision immunohistochemistry technique was employed to detect positive SCARB1 (#90332, CST, Ma, BSN, the USA) expression. Post-operative paraffin-embedded tissues were sectioned at a thickness of 4 μm and processed by following rigorous experimental protocols. A preliminary experiment was conducted to determine the optimal dilution (1:250) for formal experimentation with diaminobenzidine, P0202, Solarbio, Beijing, China) for color development. Positive controls (placenta) were included to minimize human error. SCARB1-positive staining localized in the cytoplasm, whereas Twist1-positive staining was observed in the nucleus. A 2D method was used for scoring. Staining intensity was rated as follows: No staining (0), light yellow (1), yellow (2), and brown (3). The percentage of stained cells was determined after observing three hotspots under low magnification (×100) and counting under high magnification (×200): <5% (0), 5–25% (1), 26–50% (2), 51–75% (3), and >75% (4). The final score, which ranged from 0 to 12, was determined by multiplying the two scores. A score ≤4 was considered negative, whereas a score >4 was considered positive. The positive rate was then calculated.
Synthesis of PPNs and RPPNs
PPNs were prepared by dissolving 10 mg of PLGA (P2191, Merck Millipore, Billerica, MA, the USA), 1 mg of PTX (HY-B0015, MedChemExpress, Monmouth Country, NJ, the USA), and 2 mg of tocofersolan, 57668, Merck Millipore, Billerica, MA, the USA) in 1 mL of acetone C07201102, Nanjing Reagent, Nanjing, China), followed by sonication until complete dissolution. The solution was gradually introduced into 5 mL of deionized water and stirred at 1000 rpm for 3 h at ambient temperature. The mixture was placed in a vacuum-drying oven to evaporate the acetone completely to obtain PPNs.
Blood was collected from mouse hearts and centrifuged at 3000 rpm for 5 min. The mice were administered with 1% pentobarbital sodium (21642-83-1, Shandong Xiya Chemical Industry Co., Ltd., Shandong, China) anesthesia at a dose of 40 mg/kg. Red blood cells were washed 3 times with pH 7.4 PBS to eliminate serum. The cells were resuspended in 0.2 mM ethylene diamine tetraacetic acid solution (T1300, Solarbio, Beijing, China) to induce membrane rupture. The supernatant was discarded after centrifugation at 14,800 rpm for 7 min at 4°C. This process continued until the supernatant became colorless. A total of 300 μL of RBCM suspension were combined with 2 mL of deionized water, centrifuged at 10,000 rpm for 5 min, and then resuspended. The RBCM suspension was slowly added to the PPN solution and sonicated at 100 W for 10 min. The mixture underwent 12 extrusions through a 200 nm membrane using an extruder (610000, Avanti Polar Lipids Inc., Birmingham, AL, the USA). RPPNs were obtained through these steps.
Transmission electron microscopy observation
Prussian blue, chitosan (CS)-PB@ polydopamine, and RPPNs solutions at a concentration of 1 mg/mL were placed in 1.5 mL Eppendorf tubes and thoroughly sonicated. Approximately 10 μL of each solution was dropped onto ultrathin copper grids. Excess liquid was removed with filter paper after 30 min, and the grids were air-dried. The sizes and morphological characteristics of the materials were observed using transmission electron microscopy (TEM) (Nexsa TEM, FEI Company, the USA).
Dynamic light scattering (DLS) measurement
The RBCM-PLGA-PTX-NP (RPPN) solution (1 mg/mL) was diluted 10-fold and added to a DLS cuvette. Particle size distribution (0–10 000 nm) was measured using a nanoparticle size and zeta potential analyzer (Zetasizer Ultra, Malvern Instruments, Malvern, the UK) in size mode. Data were collected and analyzed.
Zeta potential analysis
The RBCM-PLGA-PTX-NP (RPPN) solution (1 mg/mL) was diluted 10-fold and added into a cuvette. Zeta potential was measured using a nanoparticle size and zeta potential analyzer (Malvern Instruments, the UK) in zeta potential mode.
In vitro drug release experiment
In vitro, drug release behavior was investigated using a dynamic dialysis method. Nanoparticles and free PTX were placed in a dialysis bag pre-soaked in distilled water. The dialysis bag with the drug was placed in 150 mL of pH 7.4 phosphate-buffered solution in an Erlenmeyer flask. Each sample was prepared in triplicate. The temperature was kept at 37.0°C ± 0.5°C at 100 rpm. Samples (0.5 mL) were collected at pre-determined time intervals and immediately replaced with an equal volume of fresh-release medium. Cumulative release percentage was calculated, and drug release curves were plotted.
Statistical analysis
All experiments were independently repeated 3 times. Statistical analysis and graphing were performed using GraphPad Prism 8. 0 software. Comparisons between two sample means were performed using t-tests. Comparisons among multiple sample means were conducted using analysis of variance with the Student–Newman–Keuls test. Statistical significance was defined as P < 0. 05.
RESULTS
SCARB1 is overexpressed in human lung cancer tissues and associated with poor prognosis
We utilized the TCGA database to analyze SCARB1 expression characteristics and its clinical significance in lung cancer. SCARB1 was generally overexpressed in multiple tumor types, suggesting its potentially important role in the development and progression of various cancers [Figure 1a]. We analyzed SCARB1 expression in tumor and adjacent normal tissues of patients with lung cancer. Our findings revealed a significant increase in the expression of SCARB1 in tumor tissues relative to adjacent normal tissues ([Figure 1b], P < 0. 001), suggesting the potential role of SCARB1 in the initiation and progression of lung cancer. We examined the correlation between SCARB1 expression levels and lung cancer staging to understand their relationship to clinical features. Our results revealed a significant positive correlation between SCARB1 expression levels and patient clinical stages ([Figure 1c], P < 0.001). SCARB1 expression levels increased as lung cancer progressed to later stages, supporting the potential promoting role of SCARB1 in lung cancer progression. We also found a positive correlation between SCARB1 expression and lung cancer stem cell-like properties. We analyzed the correlation between SCARB1 expression and cancer stemness markers in patients with lung cancer using TCGA data from the Sangerbox 3. 0 database (http://sangerbox.com/). Elevated SCARB1 expression was significantly correlated with the upregulation of lung stemness-associated genes and enhancement of stem cell-like traits in lung adenocarcinoma tissues [Figure 1d]. Hence, SCARB1 might enhance cancer stem cell characteristics in lung cancer, possibly aiding in tumor initiation, progression, and resistance to therapy. Finally, we assessed the effect of SCARB1 expression levels on lung cancer prognosis using Kaplan–Meier survival analysis. The patients were categorized into high- and low-expression groups in accordance with the median level of SCARB1 expression. The analysis of survival curves indicated that elevated SCARB1 expression significantly reduced overall survival compared with low expression levels ([Figure 1e], log-rank test, P < 0.01). The increased expression of SCARB1 is suggested to be closely related to the poor prognosis of patients with lung cancer.
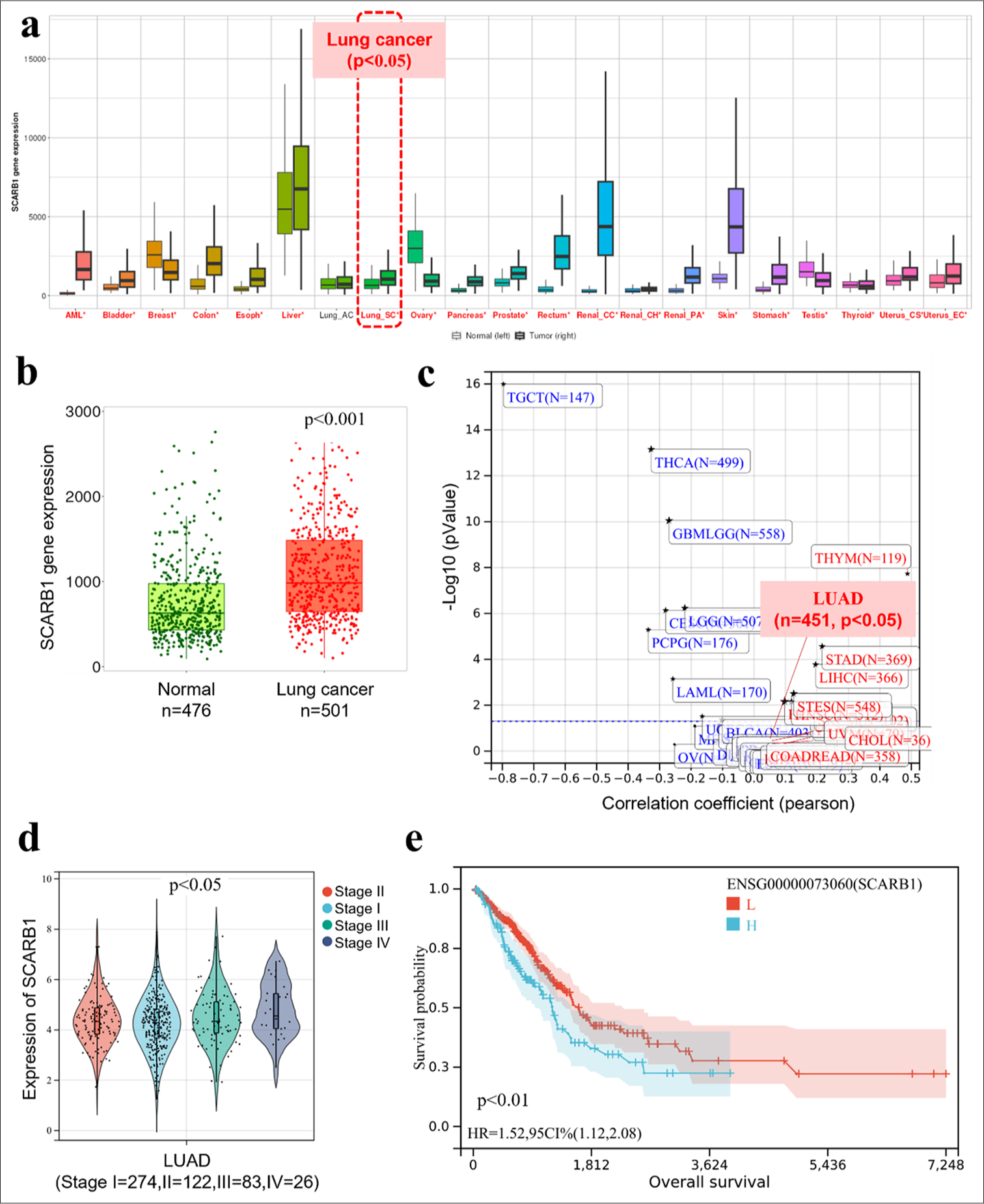
- SCARB1 is overexpressed in human lung cancer tissues and associated with poor prognosis. (a) TCGA database analysis of SCARB1 expression in tumor and adjacent normal tissues across multiple cancer types. (b) TCGA database analysis of SCARB1 expression in lung cancer tumor tissues compared with adjacent normal tissues. (c) Correlation analysis of SCARB1 expression levels with lung cancer stages. (d) Positive correlation between SCARB1 expression and stem cell-like properties in lung cancer. (e) Kaplan–Meier survival curve showing the overall survival of patients with lung cancer stratified by SCARB1 expression levels. SCARB1: Scavenger receptor class B member 1, TCGA: The Cancer Genome Atlas, LUAD: Lung adenocarcinoma.
Silencing SCARB1 inhibits lung cancer cell growth
We utilized RNA interference to suppress SCARB1 expression in A549 lung cancer cells to study its effect on cell growth. First, we designed two different siRNA sequences (si-SCARB1#1 and si-SCARB1#2) targeting SCARB1, with an empty vector (si-NC) as a control. Quantitative PCR (qPCR) and Western blot analysis results confirmed that siRNA sequences significantly decreased the messenger ribonucleic acid (mRNA) and protein levels of SCARB1 (P < 0.001). Specifically, qPCR results indicated that SCARB1 mRNA levels were significantly reduced in the si-SCARB1#1 and si-SCARB1#2 groups. Western blot analysis further confirmed significant downregulation at the protein level. SCARB1 protein expression in the si-SCARB1#1 and si-SCARB1#2 groups was significantly downregulated relative to that in the control group [Figure 2a-c]. We utilized multiple techniques to assess the effect of SCARB1 knockdown on A549 cell proliferation, colony formation, invasion, and migration. The EdU incorporation assay was employed to assess cellular DNA synthesis, indicating cell proliferation. The si-SCARB1#1 and si-SCARB1#2 groups exhibited a significant reduction in EdU-positive cells compared with the control group (P < 0.01). Hence, SCARB1 knockdown markedly inhibited A549 cell proliferation [Figure 2d and e]. Colony formation assays further validated the effect of SCARB1 knockdown on the long-term proliferation of A549 cells. The si-SCARB1#1 and si-SCARB1#2 groups exhibited a significant reduction in colony numbers compared with the control group (P < 0.01 and P < 0.001, respectively), indicating that SCARB1 knockdown impaired A549 cell proliferation and survival [Figure 2f and g]. Finally, we assessed the effect of SCARB1 knockdown on A549 cell invasion and migration using Transwell assays. Cell invasion and migration had reduced in the si-SCARB1#1 and si-SCARB1#2 groups compared with those in the control group (P < 0. 01). Hence, SCARB1 knockdown significantly inhibited A549 cell invasion and migration [Figures 2h-j]. Our experiments show that in A549 lung cancer cells, SCARB1 knockdown significantly reduced proliferation, colony formation, invasion, and migration abilities. The results confirm the hypothesis that SCARB1 substantially influences lung cancer cell proliferation and metastasis and offer substantial experimental evidence for investigating SCARB1 as a potential therapeutic target.
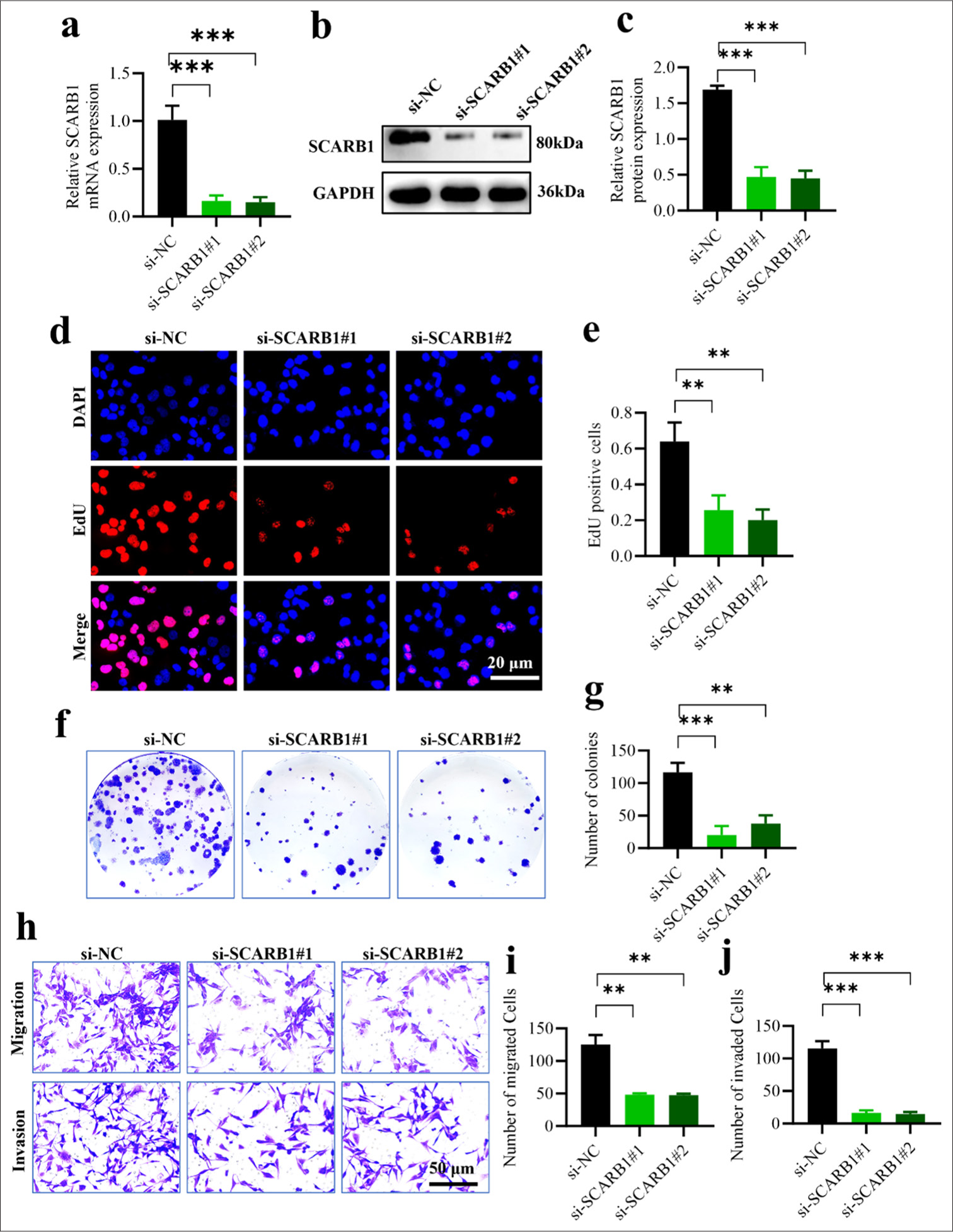
- Silencing SCARB1 inhibits lung cancer cell growth. (a) qPCR validation of SCARB1 knockdown efficiency in A549 cells. (b and c). Western blot validation of SCARB1 knockdown efficiency in A549 cells. (d and e). EdU assay of A549 cell proliferation after SCARB1 knockdown. (f and g) Colony formation assay of A549 cell growth after SCARB1 knockdown. (h-j) Transwell assay of A549 cell invasion and migration after SCARB1 knockdown. n = 3. ✶✶P < 0.01, ✶✶✶P < 0.001. SCARB1: Scavenger receptor class B member 1, EdU: 5-ethynyl-2'-deoxyuridine, DAPI: 4',6-diamidino-2'-phenylindole, GAPDH: glyceraldehyde-3-phosphate dehydrogenase, qPCR: Quantitative PCR.
Overexpression of SCARB1 enhances lung cancer cell proliferation and migration by activating Twist1
We induced SCARB1 OE in the A549 lung cancer cell line to confirm the role of SCARB1 in enhancing lung cancer cell proliferation and metastasis. First, we verified SCARB1 OE efficiency through qPCR and Western blot analyses. qPCR analysis revealed a significant 4-fold increase in SCARB1 mRNA expression in the OE-SCARB1 group compared with that in the control group (OE-NC) (P < 0.001) [Figure 3a]. Western blot analysis results confirmed a significant upregulation at the protein level; in particular, SCARB1 protein expression in the OESCARB1 group was approximately 4-fold higher than that in the control group. We also observed a 3-fold increase in Twist1 protein expression concurrent with SCARB1 OE, suggesting that SCARB1 may exert its tumor-promoting effects by upregulating Twist1 [Figure 3b-d]. We utilized multiple techniques to assess the effect of SCARB1 OE on A549 cell proliferation, colony formation, invasion, and migration. The EdU incorporation assay was employed to assess cellular DNA synthesis, which indicates cell proliferation capacity. The OE-SCARB1 group exhibited a significant increase in EdU-positive cells compared with that in the control group (P < 0.01), demonstrating that SCARB1 OE markedly enhanced A549 cell proliferation [Figure 3e and f]. Colony formation assays validated the effect of SCARB1 OE on the long-term proliferation ability of A549 cells. The OE-SCARB1 group exhibited a significant increase in colony numbers compared with the control group (P < 0. 001), confirming that SCARB1 OE enhanced A549 cell proliferation and survival [Figure 3g and h]. Finally, we assessed the effect of SCARB1 OE on A549 cell invasion and migration abilities using Transwell assays. Compared with those of the cells in the control group, the invasion and migration of the cells in the OE-SCARB1 group had significantly increased, and the difference was statistically significant (P < 0.01). Hence, SCARB1 OE significantly enhanced the invasion and migration abilities of A549 cells [Figure 3i-k]. In conclusion, SCARB1 OE in A549 lung cancer cells markedly enhanced cell proliferation, colony formation, invasion, and migration. SCARB1 OE was accompanied by the upregulation of Twist1 expression, providing important clues for the role of SCARB1 in promoting lung cancer progression through the activation of the Twist1 signaling pathway. These findings reinforce the essential role of SCARB1 in lung cancer cell proliferation and metastasis and establish a basis for detailed investigation into the molecular mechanisms of the SCARB1–Twist1 axis in lung cancer progression.
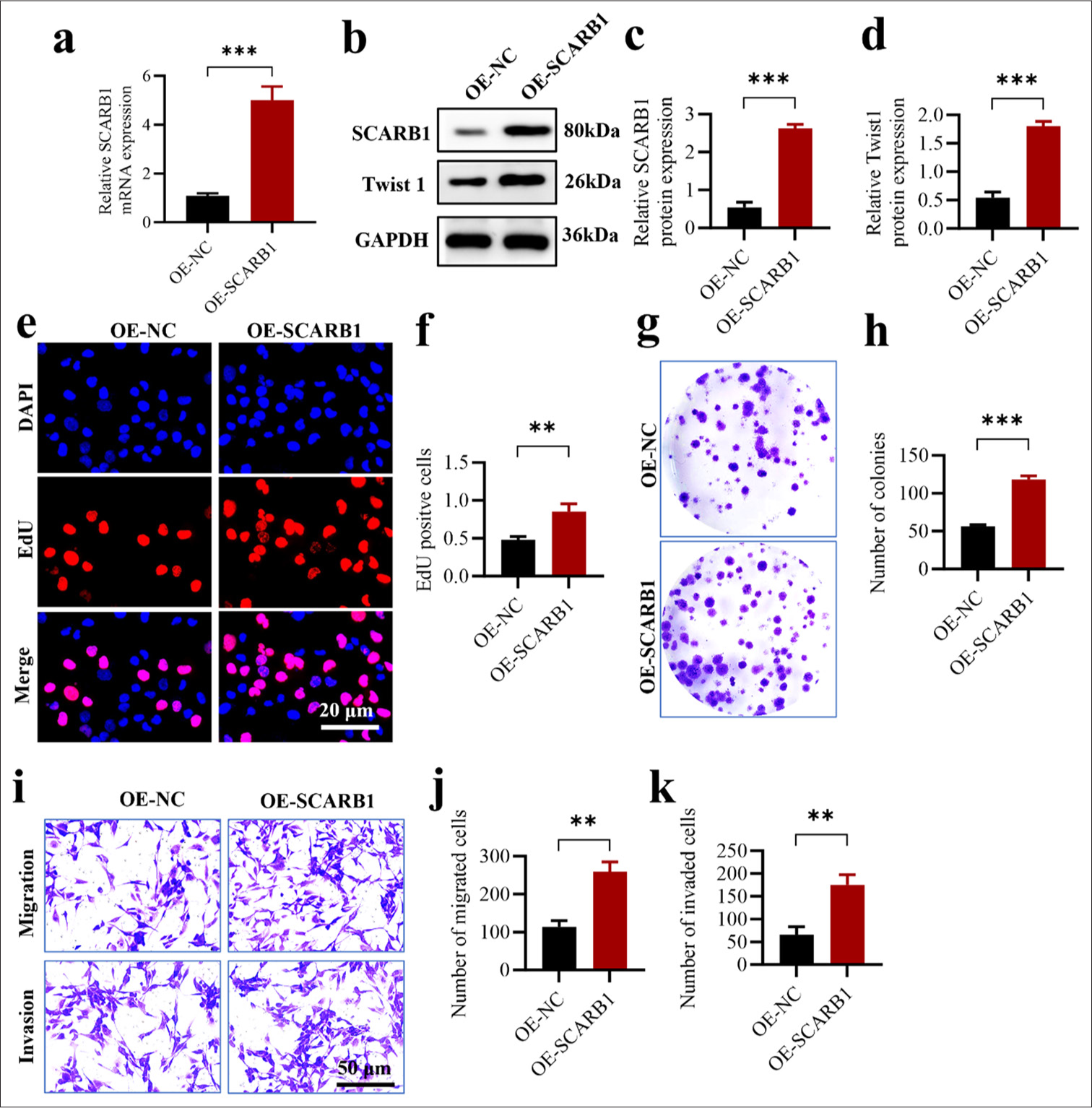
- SCARB1 overexpression enhances lung cancer cell proliferation and migration by activating Twist1. (a) qPCR validation of SCARB1 overexpression efficiency in A549 cells. (b-d) Validation of SCARB1 overexpression efficiency and Twist1 upregulation in A549 cells through Western blot analysis. (e and f) EdU assay of A549 cell proliferation after SCARB1 overexpression. (g and h) Colony formation assay of A549 cell growth after SCARB1 overexpression. (i-k) Transwell assay of A549 cell invasion and migration after SCARB1 overexpression. n = 3. ✶✶P < 0.01, ✶✶✶P < 0.001. Twist1: Twist family bHLH transcription factor 1, SCARB1: Scavenger receptor class B member 1, EdU: 5-ethynyl-2'-deoxyuridine, qPCR: Quantitative PCR.
SCARB1 OE promotes lung cancer EMT and metastasis through Twist1 activation
We developed a lung metastasis model through the tail vein injection of tumor cells to confirm the role of SCARB1 in enhancing lung cancer metastasis and explore its mechanisms. The study discovered a significant increase in lung metastatic foci in the SCARB1 OE group compared with that in the control group (P < 0.001) [Figure 4a and b]. Hence, SCARB1 has a crucial role in facilitating lung cancer metastasis. We conducted a series of IHC staining analyses on lung metastatic foci to gain deep insights into the molecular mechanisms by which SCARB1 promotes metastasis. SCARB1 IHC staining revealed significantly higher intensity in the SCARB1 OE group than in the control group (P < 0.01), confirming sustained high SCARB1 expression in metastatic foci [Figure 4c and d]. The IHC staining intensity of Twist1 in the SCARB1 OE group was also significantly enhanced (P < 0.01) relative to that in the in vitro groups, thereby supporting the hypothesis that SCARB1 promotes lung cancer metastasis by upregulating Twist1 [Figure 4e and f]. We evaluated the effect of SCARB1 OE on EMT by analyzing the expression levels of the EMT markers E-cadherin and vimentin. The SCARB1 OE group exhibited a significant decrease in the IHC staining intensity of E-cadherin (P < 0.001) and a significant increase in that of vimentin (P < 0. 001) [Figure 4g-j]. These results clearly indicate that SCARB1 OE promoted EMT in lung cancer cells. Our in vitro experiments confirm that SCARB1 enhanced lung cancer EMT and metastasis by activating the Twist1 signaling pathway. SCARB1 OE not only increased the number of lung metastatic foci but also upregulated Twist1 expression in metastatic foci; these changes were accompanied by changes in the expression pattern of typical EMT markers. The present study provides robust in vivo evidence that highlights the pivotal role of SCARB1 in lung cancer metastasis and underscores the significance of the SCARB1–Twist1 axis in modulating lung cancer EMT and metastasis.
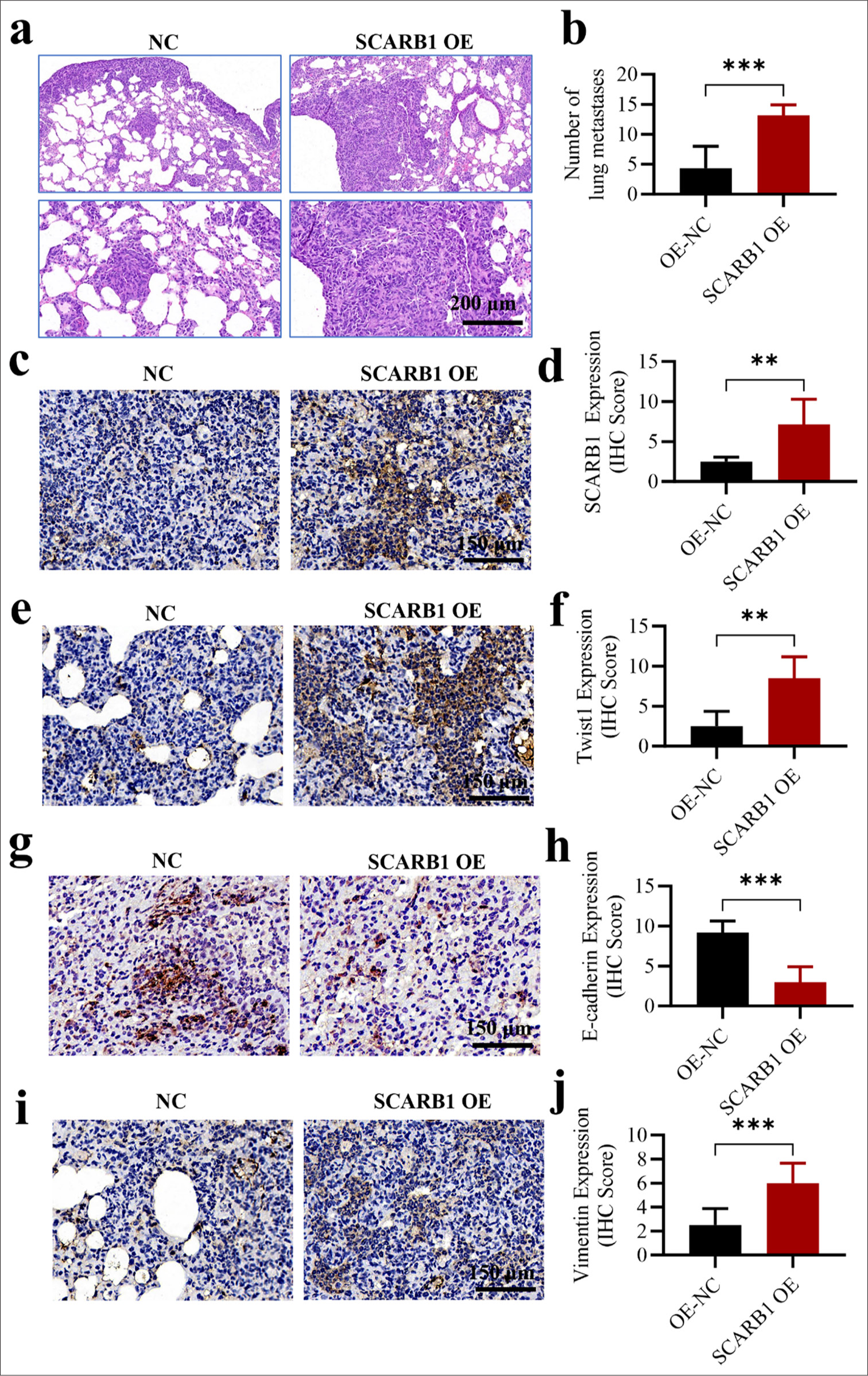
- SCARB1 overexpression promotes lung cancer EMT and metastasis through Twist1 activation in vivo. (a and b) H&E staining, objective: 100×, showed that SCARB1 overexpression enhanced the formation of metastatic nodules in the lung metastasis model after tail vein injection. (c and d) Immunohistochemical staining, Objective: 200×, of SCARB1 in metastatic tumor tissues. (e and f) Immunohistochemical staining, Objective: 200×, of Twist1 in metastatic tumor tissues. (g and h) Immunohistochemical staining, objective: 200×, of E-cadherin in metastatic tumor tissues. (i and j) Immunohistochemical staining, objective: 200×, of vimentin in metastatic tumor tissues. n = 6. ✶✶P < 0.01, ✶✶✶P < 0.001. EMT: Epithelial–mesenchymal transition, H&E: Hematoxylin and Eosin, Twist1: Twist family bHLH transcription factor 1, SCARB1: Scavenger receptor class B member 1; H&E, hematoxylin & eosin.
RPPNs inhibit SCARB1 expression in lung cancer with enhanced suppressive effects
We developed a novel nanodrug delivery system, namely RPPNs, to enhance the inhibitory effect of PTX on SCARB1. Figure 5a depicts the preparation of RPPNs and the detailed synthesis of PPNs followed by coating with RBCMs. TEM observations [Figure 5b] revealed that PPNs and RPPNs exhibited spherical structures with diameters of 100–150 nm. DLS measurements further confirmed the hydrodynamic sizes of the nanoparticles [Figure 5c], indicating that RPPNs were slightly larger than PPNs likely due to the RBCM coating. Zeta potential analysis [Figure 5d] showed that compared with PPNs, RPPNs possessed a surface charge closer to the physiological environment. This characteristic may contribute to their improved in vivo stability and biocompatibility. In vitro drug release experiments [Figure 5e-g] were performed to assess the release behavior of PTX from PPNs and RPPNs. Compared with PPNs, RPPNs exhibited a more sustained and slower drug release pattern, potentially contributing to prolonged drug action and enhanced therapeutic efficacy. We conducted a Western blot analysis to assess the effect of RPPNs on the expression levels of SCARB1 and Twist1 [Figure 5h-j]. PPNs significantly decreased the expression levels of SCARB1 and Twist1 compared with the control treatment (P < 0.01). RPPNs demonstrated superior inhibitory effects (P < 0.001), with SCARB1 and Twist1 protein levels significantly reducing. This finding confirms the superiority of RPPNs in suppressing the SCARB1– Twist1 axis. EdU cell proliferation assays [Figure 5k and l] further validated the antitumor effects of RPPNs. The PPN-treated group demonstrated a significant decrease in EdU-positive cells compared with the control group (P < 0.001), and the RPPN-treated group showed even greater inhibition of proliferation (P < 0.001). This result is consistent with the findings of Western blot analysis, further supporting the hypothesis that RPPNs inhibit tumor cell proliferation by suppressing the SCARB1–Twist1 axis. Finally, Transwell assays [Figure 5m and n] were conducted to assess the effect of RPPNs on the invasion ability of tumor cells. Their results indicated that PPNs and RPPNs significantly decreased cell invasion capability compared with the control treatment, with RPPNs exhibiting a more pronounced inhibitory effect than PPNs (P < 0.001). This finding confirms the advantage of RPPNs in suppressing tumor metastatic potential. In conclusion, we developed RPPNs, a novel nano drug delivery system with optimal physicochemical properties that effectively inhibits the SCARB1–Twist1 axis and remarkably suppresses lung cancer cell proliferation and invasion. Our results provide strong experimental evidence for the use of RPPNs as a potential therapeutic strategy for lung cancer.
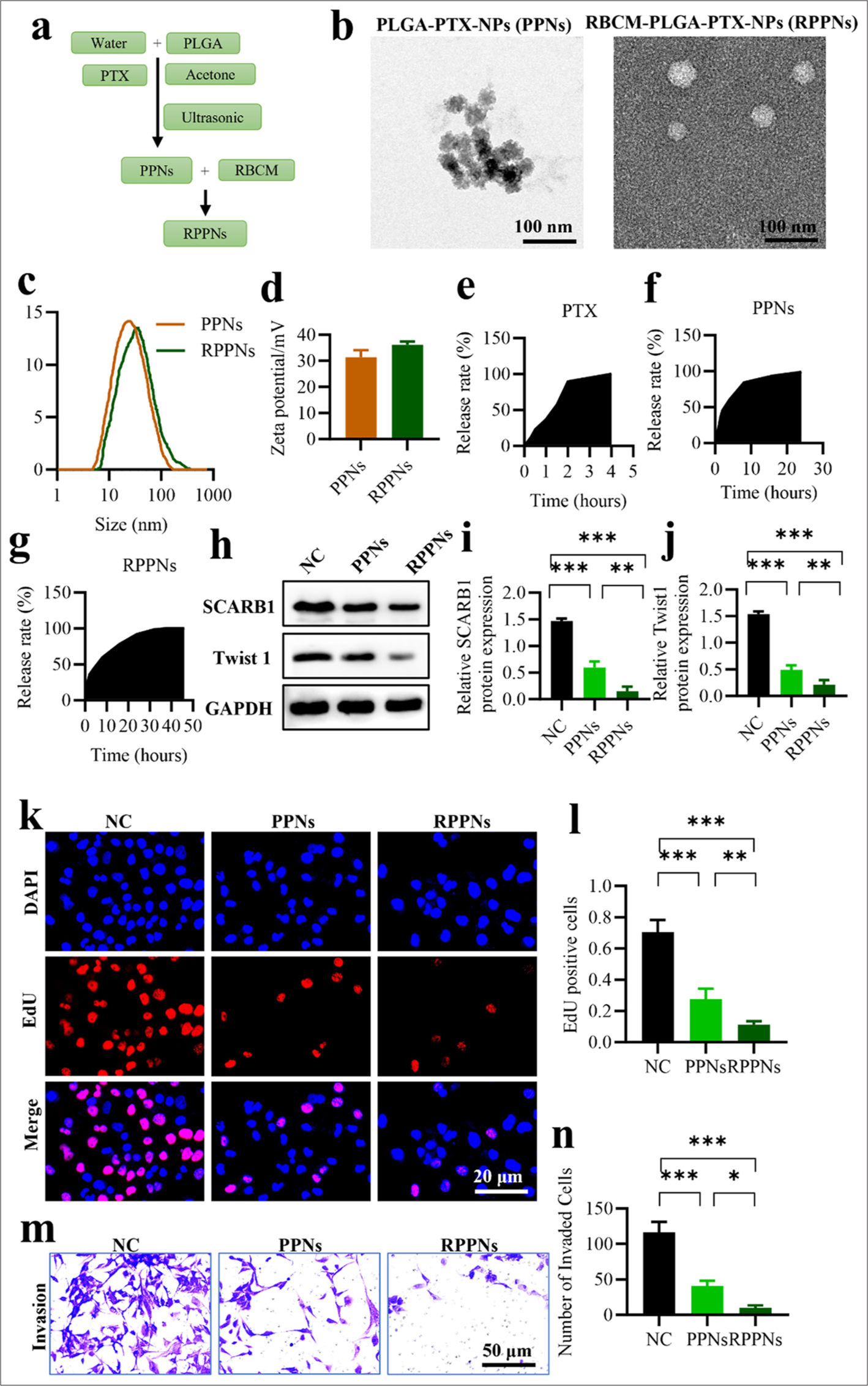
- RPPNs inhibit SCARB1 expression in lung cancer with enhanced efficacy. (a) Schematic of RPPN preparation. (b) TEM images of PPNs and RPPNs. (c) Hydrodynamic size distribution of PPNs and RPPNs. (d) Zeta potential of PPNs and RPPNs. (e-g) In vitro release profiles of PTX from PPNs and RPPNs. (h-j) Western blot analysis of SCARB1 and Twist1 protein levels in A549 cells treated with PPNs or RPPNs. (k and l). EdU assay of A549 cell proliferation after treatment with PPNs or RPPNs. (m and n) Transwell assay of A549 cell invasion after treatment with PPNs or RPPNs. n = 3. ✶P < 0.05, ✶✶P< 0.01, ✶✶✶P < 0.001. RPPNs, red blood cell membrane-coated poly (lactic-co-glycolic acid) nanocarriers loaded with paclitaxel; PPNs, poly (lactic-co-glycolic acid) nanocarriers loaded with paclitaxel; PTX: Paclitaxel, Twist1: Twist family bHLH transcription factor 1, SCARB1: Scavenger receptor class B member 1, EdU: 5-ethynyl-2'-deoxyuridine, TEM: Transmission electron microscopy.
SCARB1 OE antagonizes the antimetastatic effects of RPPNs
To further validate the crucial role of SCARB1 in lung cancer metastasis and the therapeutic efficacy of RPPNs, we designed an in vivo experiment comprising four groups: The control, RPPNs treatment, SCARB1 OE, and RPPNs + SCARB1 OE groups. A lung metastasis model was developed by injecting tumor cells into the tail veins of mice. We found that RPPN treatment significantly decreased lung metastatic foci compared with the control treatment (P < 0.001), whereas SCARB1 OE significantly increased the number of metastatic foci (P < 0.001). In the RPPNs + SCARB1 OE group, SCARB1 OE partially negated the antimetastatic effect of RPPNs, resulting in a number of metastatic foci comparable with those in the control group [Figure 6a and b]. This result strongly supports the promoting role of SCARB1 in lung cancer metastasis while also confirming the mechanism by which RPPNs inhibit lung cancer metastasis by targeting SCARB1. We performed qRT-PCR analysis on metastatic tissue samples to gain deep insights into the molecular mechanisms of RPPNs. SCARB1 mRNA expression was notably reduced in the RPPN treatment group (P < 0. 001) and markedly elevated in the SCARB1 OE group (P < 0. 001). The SCARB1 expression level in the RPPNs + SCARB1 OE group resembled that in the control group, indicating that SCARB1 OE can partially mitigate the effect of RPPNs [Figure 6c]. The mRNA expression patterns of Twist1 were similar to those of SCARB1, supporting the hypothesis that SCARB1 promotes lung cancer metastasis through the Twist1 signaling pathway [Figure 6d]. This finding also explains why SCARB1 OE could antagonize the antimetastatic effects of RPPNs. We evaluated the mRNA expression levels of E-cadherin and vimentin to analyze changes in EMT. E-cadherin expression notably increased in the RPPN treatment group and significantly decreased in the SCARB1 OE group (P < 0.001, [Figure 6e]). Vimentin, a mesenchymal marker, notably decreased in the RPPN treatment group (P < 0. 01) and markedly increased in the OE-SCARB1 group (P < 0. 01) [Figure 6f]. E-cadherin and Vimentin expression levels in the RPPNs + SCARB1 OE group were similar to those in the control group, indicating that SCARB1 OE could counteract the inhibitory effects of RPPNs on EMT. In conclusion, these results strongly confirm the critical role of SCARB1 in promoting lung cancer metastasis and its mechanism of regulating EMT through the Twist1 signaling pathway. Our study validates RPPNs as an effective therapeutic strategy capable of inhibiting lung cancer metastasis by targeting the SCARB1–Twist1 axis. However, SCARB1 OE can partially counteract the therapeutic effects of RPPNs, providing important evidence for developing effective combination therapy strategies in the future.
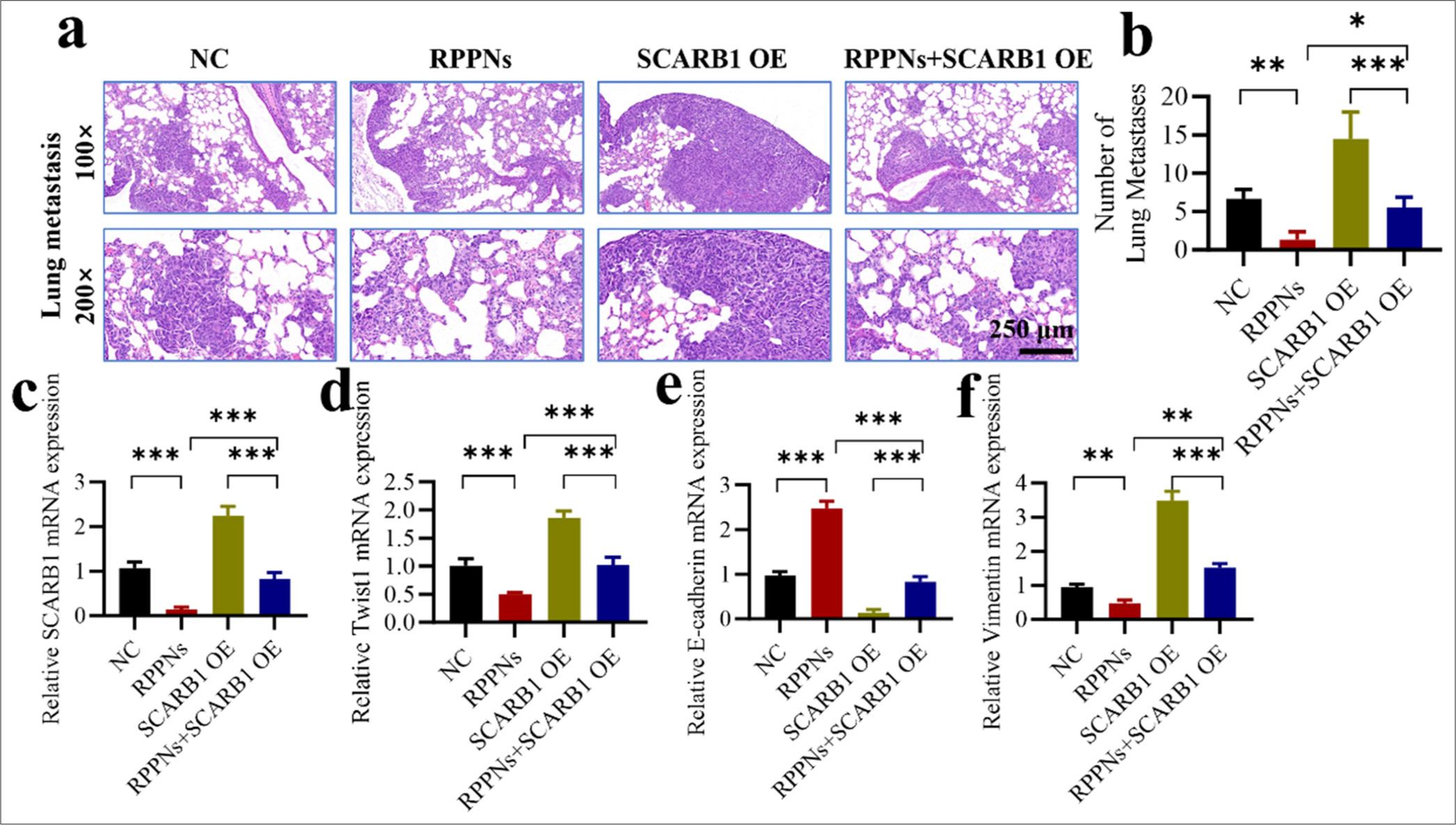
- SCARB1 overexpression antagonizes the antimetastatic effects of RPPNs. (a and b) The lung metastasis model established through tail vein injection demonstrates that SCARB1 overexpression antagonizes the antitumor effects of RPPNs. (c) qRT-PCR analysis of SCARB1 expression in metastatic tumor tissues. (d) qRT-PCR analysis of Twist1 expression in metastatic tumor tissues. (e) qRT-PCR analysis of E-cadherin expression in metastatic tumor tissues. (f) qRT-PCR analysis of Vimentin expression in metastatic tumor tissues. n = 6. ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. qRT-PCR: Quantitative real-time polymerase chain reaction, Twist1: Twist family bHLH transcription factor 1, SCARB1: Scavenger receptor class B member 1.
DISCUSSION
This study explored the role and molecular mechanisms of SCARB1 in lung cancer progression and developed a novel nanoparticle drug delivery system targeting SCARB1. Our findings reveal that SCARB1 promotes lung cancer growth and metastasis by activating the Twist1 signaling pathway and demonstrates the efficacy of RPPNs as a promising therapeutic strategy.
We discovered that SCARB1 is overexpressed in human lung cancer tissues and is closely associated with poor patient prognosis. This finding is consistent with the results of previous studies on other cancers. For example, GonzaloCalvo et al. reported that SCARB1 is overexpressed in breast cancer and correlated with tumor invasiveness and poor patient outcomes.[11] This study is the first to validate the association of SCARB1 with lung cancer, suggesting its potential as a prognostic marker.
Through in vitro experiments, we demonstrated that silencing SCARB1 remarkably inhibits the growth, invasion, and migration capabilities of lung cancer cells. Our study highlights SCARB1’s functional importance across various tumors, underscoring its potential as a therapeutic target.[25,26] We identified the molecular mechanism by which SCARB1 enhances lung cancer cell proliferation, migration, and EMT through the activation of the Twist1 signaling pathway. Twist1 is a pivotal EMT regulator, considerably influencing tumor invasion and metastasis.[27,28] Our study not only confirms the role of Twist1 in lung cancer progression but also elucidates the regulatory relationship between SCARB1 and Twist1 for the 1st time. This finding offers a novel insight into the molecular mechanisms of lung cancer metastasis and establishes a basis for creating therapies targeting the SCARB1–Twist1 axis.
In vivo experiments involved creating a lung metastasis model through the tail vein injection of tumor cells and further confirmed the role of SCARB1 in enhancing lung cancer metastasis. SCARB1 OE markedly elevated lung metastatic foci in correlation with increased Twist1 expression and vimentin levels and decreased E-cadherin. These results not only support the findings of our in vitro experiments but also highlight the critical role of the SCARB1–Twist1 axis in lung cancer metastasis.
On the basis of these findings, we developed a novel SCARB1-targeted nanoparticle drug delivery system, namely RPPNs. This nanocarrier not only effectively loads PTX but also exhibits excellent biocompatibility and targeting ability due to its RBCM coating. Our findings indicate that RPPNs more effectively inhibit SCARB1 expression and suppress lung cancer cell proliferation and invasion than traditional PPNs. This result is consistent with the excellent performance of the RBCM-coated nanocarriers developed for breast cancer treatment,[29,30] further confirming the potential of the RBCM coating strategy in enhancing the efficacy of nanoparticle drug delivery systems.[31] In addition, the release time of RPPNs in the body is long, which, combined with previous reports, may be the result of strengthened particle stability, improved rigidity, and increased material encapsulation reliability.[32] Through the nanostructure of RBCM, the residence time of drugs in the body is improved, providing another delivery system for the biomedical field. SCARB1 OE partially antagonizes the antimetastatic effects of RPPNs. This finding underscores the importance of SCARB1 as a therapeutic target for RPPNs and highlights the necessity of considering SCARB1 expression levels in tumor cells for assessing treatment efficacy in future clinical applications.
While this study presents crucial findings, certain limitations remain, which warrant further investigation in future research. First, while we have demonstrated that SCARB1 promotes lung cancer progression by activating Twist1, the specific molecular mechanism by which SCARB1 regulates Twist1 remains unclear. Future studies could employ protein interaction analysis, transcriptome sequencing, and other methods to delve deeply into the regulatory network between SCARB1 and Twist1. Second, although RPPNs have shown good antitumor effects in vitro and animal experiments, their safety and efficacy in clinical applications need further evaluation. In consideration of tumor heterogeneity, future research should explore the quantitative relationship between SCARB1 expression levels and RPPN treatment efficacy to provide a basis for personalized therapy.
SUMMARY
This study reveals the molecular mechanism by which SCARB1 enhances lung cancer growth and metastasis through the Twist1 signaling pathway and introduces a novel SCARB1-targeted nanoparticle drug delivery system, namely RPPNs. Its findings enhance our comprehension of the molecular mechanisms promoting lung cancer progression and offer novel strategies for the diagnosis and treatment of lung cancer. In the future, further research should be conducted on the regulatory mechanisms of the SCARB1– Twist1 axis and the clinical translation of RPPNs. We hope to provide treatment options with increased precision and effectiveness to patients with lung cancer, ultimately improving their survival rates and quality of life.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study were available from the corresponding author on reasonable request.
ABBREVIATIONS
DLS: Dynamic light scattering
EdU: 5-ethynyl-2’-deoxyuridine
EMT: Epithelial–mesenchymal transition
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
PBS: Phosphate buffer saline
PLGA: Poly (lactic-co-glycolic acid)
PTX: Paclitaxel
qRT-PCR: Quantitative real-time polymerase chain reaction
RBCM: Red blood cell membrane
RBCM-PLGA-PTX-NPs or RPPNs: PTX-loaded PLGA nanocarriers coated with RBCM
SCARB1: Scavenger receptor class B member 1
SDS-PAGE: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
SPF: Specific pathogen-free
TCGA: The Cancer Genome Atlas
TEM: Transmission electron microscopy
AUTHOR CONTRIBUTIONS
XL and HB: Designed the study; all authors conducted the study; TXD and YS: Collected and analyzed the data; XL and HB: Participated in drafting the manuscript, and all authors contributed to the critical revision of the manuscript for important intellectual content. All authors gave final approval of the version to be published. All authors participated fully in the work, took public responsibility for appropriate portions of the content, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or completeness of any part of the work were appropriately investigated and resolved.
ACKNOWLEDGMENT
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Shenzhen Ling Fu Tuopu Biotechnology Co., LTD. The study was approved by the Institutional Animal Care and Use Committee of Shenzhen Ling Fu Tuopu Biotechnology Co., LTD. (TOP-IACUC-2024-0189). Ethics date. July 26, 2024. This article does not involve human experiments and does not require informed consent from patients.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: This work was supported by the Shenzhen Nanshan District Health System Science and Technology Major Project (Innovative research projects between District and university) (No. NSZD2024063).
References
- Lung cancer pathology: Current concepts. Clin Chest Med. 2020;41:67-85.
- [CrossRef] [PubMed] [Google Scholar]
- The global burden of lung cancer: Current status and future trends. Nat Rev Clin Oncol. 2023;20:624-39.
- [CrossRef] [PubMed] [Google Scholar]
- Lung cancer LDCT screening and mortality reduction-evidence, pitfalls and future perspectives. Nat Rev Clin Oncol. 2021;18:135-51.
- [CrossRef] [PubMed] [Google Scholar]
- Screening for lung cancer: 2023 guideline update from the American Cancer Society. CA Cancer J Clin. 2024;74:50-81.
- [CrossRef] [PubMed] [Google Scholar]
- Targeted therapy in advanced non-small cell lung cancer: Current advances and future trends. J Hematol Oncol. 2021;14:1-20.
- [CrossRef] [PubMed] [Google Scholar]
- Targeted therapies for lung cancer patients with oncogenic driver molecular alterations. J Clin Oncol. 2022;40:611-25.
- [CrossRef] [PubMed] [Google Scholar]
- High HDL-cholesterol paradox: SCARB1-LAG3-HDL axis. Curr Atheroscler Rep. 2021;23:1-6.
- [CrossRef] [PubMed] [Google Scholar]
- High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc Natl Acad Sci. 2017;114:5219-24.
- [CrossRef] [PubMed] [Google Scholar]
- Selected single-nucleotide polymorphisms in FOXE1, SERPINA5, FTO, EVPL, TICAM1 and SCARB1 are associated with papillary and follicular thyroid cancer risk: Replication study in a German population. Carcinogenesis. 2016;37:677-84.
- [CrossRef] [PubMed] [Google Scholar]
- SCARB1 in extracellular vesicles promotes NPC metastasis by co-regulating M1 and M2 macrophage function. Cell Death Discov. 2023;9:323.
- [CrossRef] [PubMed] [Google Scholar]
- Intratumor cholesteryl ester accumulation is associated with human breast cancer proliferation and aggressive potential: A molecular and clinicopathological study. BMC Cancer. 2015;15:1-14.
- [CrossRef] [PubMed] [Google Scholar]
- Establishment and characterization of a highly metastatic human osteosarcoma cell line from osteosarcoma lung metastases. J Bone Oncol. 2021;29:100378.
- [CrossRef] [PubMed] [Google Scholar]
- TWIST1 correlates with Notch signaling pathway to develop esophageal squamous cell carcinoma. Mol Cell Biochem. 2020;474:181-8.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular mechanisms of TWIST1?regulated transcription in EMT and cancer metastasis. EMBO Rep. 2023;24:e56902.
- [CrossRef] [PubMed] [Google Scholar]
- Loss of FBXL14 promotes mesenchymal shift and radioresistance of non-small cell lung cancer by TWIST1 stabilization. Signal Transduct Target Ther. 2021;6:272.
- [CrossRef] [PubMed] [Google Scholar]
- TWIST1 is a critical downstream target of the HGF/MET pathway and is required for MET driven acquired resistance in oncogene driven lung cancer. Oncogene. 2024;43:1431-4.
- [CrossRef] [PubMed] [Google Scholar]
- Perspectives of nano-carrier drug delivery systems to overcome cancer drug resistance in the clinics. Cancer Drug Resist. 2021;4:44.
- [CrossRef] [PubMed] [Google Scholar]
- Nanocarrier-based drug delivery system for cancer therapeutics: A review of the last decade. Curr Med Chem. 2021;28:3753-72.
- [CrossRef] [PubMed] [Google Scholar]
- Recent advances of multifunctional PLGA nanocarriers in the management of triple-negative breast cancer. AAPS PharmSciTech. 2023;24:258.
- [CrossRef] [PubMed] [Google Scholar]
- Transferrin-conjugated red blood cell membrane-coated poly (lactic-co-glycolic acid) nanoparticles for the delivery of doxorubicin and methylene blue. ACS Appl Nano Mater. 2020;3:3807-19.
- [CrossRef] [Google Scholar]
- Photolytic removal of red blood cell membranes camouflaged on nanoparticles for enhanced cellular uptake and combined chemo-photodynamic inhibition of cancer cells. Mol Pharm. 2022;19:805-18.
- [CrossRef] [PubMed] [Google Scholar]
- Stealth polydopamine-based nanoparticles with red blood cell membrane for the chemo-photothermal therapy of cancer. ACS Appl Bio Mater. 2020;3:2350-9.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging nanomedicines of paclitaxel for cancer treatment. J Control Rel. 2022;342:280-94.
- [CrossRef] [PubMed] [Google Scholar]
- Co-delivery of paclitaxel (PTX) and docosahexaenoic acid (DHA) by targeting lipid nanoemulsions for cancer therapy. Drug Deliv. 2022;29:75-88.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of common genetic variants in vitamin E-related pathway genes and colorectal cancer susceptibility. Arch Toxicol. 2021;95:2523-32.
- [CrossRef] [PubMed] [Google Scholar]
- H19 recruited N 6-methyladenosine (m 6 A) reader YTHDF1 to promote SCARB1 translation and facilitate angiogenesis in gastric cancer. Chin Med J. 2023;136:1719-31.
- [CrossRef] [PubMed] [Google Scholar]
- TWIST1 activates cancer stem cell marker genes to promote epithelial-mesenchymal transition and tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer. 2022;22:1272.
- [CrossRef] [PubMed] [Google Scholar]
- The transactivation domain of TWIST1 is required for TWIST1-induced aggressiveness in non-small cell lung cancer. Cancer Res. 2020;80:6060.
- [CrossRef] [Google Scholar]
- White blood cell membrane?coated nanoparticles: Recent development and medical applications. Adv Healthc Mater. 2022;11:2101349.
- [CrossRef] [PubMed] [Google Scholar]
- Cell membrane coated nanocarriers-an efficient biomimetic platform for targeted therapy. J Control Release. 2020;327:546-70.
- [CrossRef] [PubMed] [Google Scholar]
- Red blood cell membrane-coated upconversion nanoparticles for pretargeted multimodality imaging of triple-negative breast cancer. Biomater Sci. 2020;8:1802-14.
- [CrossRef] [PubMed] [Google Scholar]
- Recent advances on the application of nanobiomimetic structures as drug delivery systems. J Drug Deliv Sci Technol. 2024;100:106009.
- [CrossRef] [Google Scholar]








