Translate this page into:
Suppression of regulatory factor X 7 alleviates airway remodeling and inflammation in childhood asthma

*Corresponding author: Fang Liu, Department of Pediatrics, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China. liufangsh30@163.com
-
Received: ,
Accepted: ,
How to cite this article: Wu Y, Dai T, Qin J, Guo J, Fan J, Mei J, et al. Suppression of regulatory factor X 7 alleviates airway remodeling and inflammation in childhood asthma. CytoJournal. 2025;22:15. doi: 10.25259/Cytojournal_138_2024
Abstract
Objective:
Childhood asthma is a chronic heterogeneous syndrome composed of distinct disease entities or phenotypes. This study was conducted to characterize regulatory factor X 7 (RFX7) in childhood asthma.
Material and Methods:
Two available transcriptome datasets (GSE65204 and GSE27011) were used to analyze regulatory factor X (RFX) family members in childhood asthma. Random forest, logistic regression, and linear support vector machine (SVM) analyses were performed to construct an RFX-based classification model. Airway smooth muscle cells (ASMCs) were induced through platelet-derived growth factor-BB (PDGF-BB) for an asthma in vitro model. RFX7 expression was measured through immunoblotting. RFX7 was knocked out by transfection of RFX7 small-interfering RNAs, and then airway remodeling and inflammation were assayed.
Results:
Among RFX family members, RFX3, RFX7, and RFX-associated protein displayed differential expression in childhood asthma versus healthy controls. Thus, SVM, logistic regression, and random forest-based machine learning models were built. The random forest model presented the best diagnostic efficacy (area under the curve [AUC] = 1 and 0.67 in discovery and verification sets). RFX7 was found to be effective in diagnosing childhood asthma (AUC = 0.724 and 0.775 in discovery and verification sets). In addition, RFX7 was overexpressed in PDGF-BB-stimulated ASMCs (✶✶P < 0.01). Silencing RFX7 remarkably attenuated the proliferative and migrative capacities of ASMCs with PDGF-BB stimulation (✶✶P < 0.01). In addition, RFX7 was positively related to neutrophil infiltration in childhood asthma, and its knockdown downregulated the levels of pro-inflammatory cytokines in PDGF-BB-stimulated ASMCs (✶✶P < 0.01).
Conclusion:
The findings of this study indicate that RFX7 is a novel molecule that is correlated with airway remodeling and inflammation in childhood asthma, providing insights into the mechanism underlying this disease and its potential clinical importance.
Keywords
Airway remodeling
Airway smooth muscle cells
Asthma
Inflammation
Regulatory factor X transcription factors
INTRODUCTION
Asthma remains a prevalent chronic non-communicable disease with reversible airflow obstruction, which affects approximately 350 million individuals and results in a notable socio-economic and health burden worldwide.[1] Children are more susceptible to asthma due to their immature physical development and immune systems.[2] As a dominating chronic inflammatory disease in childhood, the estimated prevalence of childhood asthma ranges from 2.6% to 30.5%,[3] which continues to increase in many regions and countries, particularly in low-and middle- income countries.[4] Several risk factors for childhood asthma have been investigated, for example, parental asthma, childhood overweight or obesity, and preterm birth.[5-7] Asthma symptoms range from mild to severe, with a multifactorial etiology that begins in the fetal period.[8] At present, mild asthma can be controlled; nonetheless, severe asthma remains an alarming burden. Improving childhood asthma control is an urgent need globally, particularly in less affluent countries.[9] Serious distortion of airway smooth muscle cells (ASMCs) represents a frequent phenomenon of severe asthma.[10] Asthmatic ASMCs play a role in hyperresponsiveness as well as inflammatory and remodeling processes.[11,12] In addition, aberrant ASMCs in asthma can result in hyperplasia, hypertrophy, and the secretion of inflammatory mediators and extracellular matrix (ECM) proteins.[13-15] Hence, the development of effective treatment regimens needs an in-depth understanding of the machinery of asthmatic ASMCs.
Childhood asthma is a highly heterogeneous syndrome composed of diverse disease entities or phenotypes, which is due to complex gene-gene and gene-environment interactions.[16] Regulatory factor X (RFX) family members share a highly conserved winged-helix DNA-binding domain that recognizes X-box DNA motifs, which were first discovered in mammals.[17] RFX transcription factors protect major histocompatibility complex class II genes against epigenetic silencing by DNA methylation.[18] One DNA microarray profiling-based study indicated that RFX genes mediate antigen processing and presentation pathways in childhood asthma.[19] RFX5 is a close relative of RFX7, and the expression of most RFX genes is limited to specific cell types. RFX1, RFX5, and RFX7 show universal expression.[20] RFX7 has emerged as a tumor suppressor,[21] which can be activated by p53 and cellular stress (DNA damage, ribosomal stress, etc.) across diverse cell types.[20] In addition, RFX7 hinders the metabolic activity of natural killer cells as well as facilitates their maintenance and immunity.[22] Nonetheless, RFX7 is uncharacterized in childhood asthma. In addressing the aforementioned limitation, the importance and potential transcriptional mechanisms in childhood asthma were systematically assessed in this study.
MATERIAL AND METHODS
Data acquisition
Microarrays of 36 atopic asthmatic and 33 non-atopic nonasthmatic children were acquired from GSE65204 of the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65204).[23] The dataset was based on the GPL14550 platform. Microarrays of 36 childhood asthma and 18 controls were also collected from GSE27011 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27011).[24,25] The microarray data were converted from logarithm to base 2. GSE65204 and GSE27011 were adopted as discovery and verification sets, respectively.
Differential expression analysis
Limma software (R 3.6.1 software) package powers differential expression analysis for microarray and RNA-sequencing data.[26] Asthmatic and non-asthmatic groups were compared using the package. Differentially expressed genes (DEGs) were selected on the basis of fold change >1.2 and P < 0.05.
Enrichment analyses of Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
Clusterprofiler package (×4) can automate biological term classification and enrichment analysis of specified genes.[27] Through running this package, GO and KEGG pathway enrichment analyses of DEGs were implemented. Terms or pathways with P < 0.05 were considered significantly enriched.
Gene set variation analysis (GSVA)
GSVA, a gene set enrichment approach (GSEA), provides enhanced power to evaluate the variation in pathway activity in an unsupervised manner.[28] Based on the RFX family members, the single–sample GSEA (ssGSEA) score of the RFX signature was estimated.
Machine learning
Scikit-learn (https://scikit-learn.org/stable/), a Python machine learning library, was adopted for random forest, logistic regression, and linear support vector machine (SVM) analyses to establish an RFX-based classification model. The performance was evaluated on the basis of the receiver operating characteristic (ROCs) curves, with subsequent estimation of the area under the ROCs.
Cell culture and treatment
Human ASMCs (CTCC-056ASMC, Pubebio, Wuxi, China) were maintained in Dulbecco’s Modified Eagle’s Medium (Solarbio, Beijing, China) composed of 10% fetal bovine serum (FBS; S9020, Solarbio, Beijing, China) with 1% penicillin/streptomycin (P7630, Solarbio, Beijing, China) in a 5% Carbon dioxide incubator (Scientific 8000, Thermo, USA) at 37°C. The culture medium was changed every three days. To establish an asthma cell model, ASMCs were administrated with 50 ng/mL platelet-derived growth factor BB (PDGF-BB; abs05159, Absin, Shanghai, China) for 24 h.[29] Cell lines were authenticated by the Genomics Unit at Clinical Laboratory Management Association (CLMA), using short tandem repeat profiling (AmpFLSTR ldentifler Plus polymerase chain reaction [PCR] Amplification Kit; Applied Biosystems 4427368). A mycoplasma test was performed in all cell lines every other week using the MycoAlert Mycoplasma Detection Kit (LT07–710, LONZA, USA).
Western blot
ASMCs were mixed with radio-immunoprecipitation assay buffer (P0013B, Beyotime, Shanghai, China) combined with protease and phosphatase inhibitors (ST505, Beyotime, Shanghai, China), with subsequent homogenization at 4°C for 30 min. Following centrifugation, supernatant samples were collected. Protein concentration was tested by utilizing a bicinchoninic acid reagent (BL521A, Biosharp, Beijing, China). A total of 20 μg of proteins was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, with subsequent transference onto nitrocellulose membranes (HATF00010, Millipore, Boston, USA). A primary antibody targeting RFX7 (1 μg/mL, Rabbit; A303–062A, Thermo Fisher Scientific, USA) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1/10000; Rabbit; ab181602; Abcam, Boston, USA) was adopted in accordance with the manufacturer’s specifications. Horseradish peroxidaseconjugated goat anti-rabbit immunoglobulin G (IgG) (1/5000; ab97051; Abcam, Boston, USA) was utilized as the secondary antibody. Enhanced chemiluminescence reagent (ECL) (Millipore, Bedford, MA) was used to visualize protein bands. Such protein bands were developed on a ChemiDoc XRS system (Bio-rad, Hercules, USA). Quantitative analyses were conducted using ImageJ software (National Institutes of Health, Maryland, USA).
Transient transfection
Small interfering RNAs (siRNAs) against RFX7 (which is known as small interfering RFX7 [si-RFX7]) and their controls (small interfering-negative control [si-NC]) were synthesized on the basis of GenePharma (Shanghai, China). The transfection sequence is presented as follows: (si-RFX7-1: AGUUCUUGAUCUUGUGUUGCA CAACACAAGAUCAAGAACUCC; siRFX7-2: UUAGAUUGUACAGUUUUGCAG GCAAAACUGUACAAUCUAAAG; siRFX7-3: UUUAGAUUGUACAGUUUUGCA CAAAACUGUACAAUCUAAAGU; siRNANC: UUCUCCGAACGUGUCACGUTT ACGUGACACGUUCGGAGAATT). ASMCs were seeded onto six-well plates (2 × 105 cells/mL). When confluence reached 80%, transient transfection was conducted by using Lipofectamine 3000 (L3000150, Invitrogen, Carlsbad, USA) in accordance with the manufacturer’s procedures. ASMCs were gathered following 48-h transfection for subsequent experiments.
5-Ethyny-2'Edeoxyuridine (EdU) assay
ASMC proliferation was evaluated using the EdU assay. ASMCs were seeded onto a 24-well plate. Subsequently, they were exposed to a fresh medium supplemented with 50 nM EdU reagent (C0075S, Beyotime, Shanghai, China) for 2 h at 37°C, with subsequent fixation by 4% paraformaldehyde (P0099, Beyotime, Shanghai, China). After staining with Hoechst 33258 (C1017, Beyotime, Shanghai, China), ASMCs were subjected to fluorescence microscopy (IX73, OLYMPUS, Germany).
Wound healing assay
ASMCs were inoculated into six-well plates (2 × 106/well) and then mechanically disrupted utilizing a sterile 200 μL pipette tip, thereby creating a linear wound. After rinsing with phosphate-buffered saline, the wells were filled with a culture medium without FBS and then further incubated for 48 h. Under a light microscope (Zeiss), photographs were acquired at appropriate times, and closure ratios were estimated using ImageJ software.
Transwell assay
Transwell filter chambers with an 8-μm pore (Corning, New York, USA) were adopted for migration evaluation. ASMCs (4 × 105 cells/mL) in a serum-free medium were planted onto the upper well of the chamber, with 500 μL of culture medium supplemented with 10% FBS added to the lower well. Subsequently, cells were continuously incubated for 24 h. Then, migrated cells were fixed in 4% paraformaldehyde (Beyotime) for 15 min and dyed with 0.1% crystal violet reagent (Beyotime) for 15 min. Furthermore, the migrated cells were photographed by using a light microscope (XDS-1A, Shanghai Precision Scientific Instrument Co., LTD., China). The data were analyzed using ImageJ software.
Immunofluorescent staining
ASMCs were fixed in 4% paraformaldehyde for 15 min, with subsequent permeabilization with 0.3% Triton X-100 (P0096, Beyotime, Shanghai, China). Following administration with 5% bovine serum albumin buffer (ST2249, Beyotime, Shanghai, China), ASMCs were incubated with a primary antibody against alpha-smooth muscle actin (α-SMA; 1/250; ab124964; Abcam, Boston, USA), N-cadherin (1/250; ab18203; Abcam; Boston, USA), or E-cadherin (1/250; ab40772; Abcam; Boston, USA) overnight at 4 °C. Afterward, they were exposed to goat anti-rabbit IgG H&L Alexa Fluor 488 (1/200; ab150077; Abcam; Boston, USA) or Alexa Fluor 647 (1/200; ab150079; Abcam; Boston, USA) for 1 h at room temperature. Subsequently, coverslips were dyed by Hoechst 33258. Slides were mounted and detected using a fluorescence microscope (OLYMPUS IX73, Japan).
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA extraction was achieved using TRIzol (15596018CN, Invitrogen, USA) in accordance with the manufacturer’s procedures. Complementary DNA was synthesized using the PrimeScript RT reagent Kit (K1622, Thermo, USA). Subsequently, RT-qPCR was implemented using synergetic binding reagent (SYBR) Premix Ex Taq II (F-415XL, Thermo, USA) on ABI (real-time fluorescence quantitative PCR instrument) 7500 Real-Time PCR System (ABI7500, ThermoFisher, Waltham, USA). The primers used were as follows: tumor necrosis factor alpha (TNF-α), 5'-TCTTCTCCTTCCTGATCGT-3' (sense), 5'-GCTACAGGCTTGTCACTC-3' (antisense); interleukin (IL)-1 beta, 5'-ATGACCTGAGCACCTTCT-3' (sense), 5'-GGACCAGACATCACCAAG-3' (antisense); IL-6 (IL-6), 5'-TGAGAGTAGTGAGGAACAAG-3' (sense), 5'-CGCAGAATGAGATGAGTTG-3' (antisense); GAPDH, 5'-CATCACCATCTTCCAGGAG-3' (sense), 5'-AGGCTGTTGTCATACTTCTC-3' (antisense). Relative expression values were estimated using the 2−ΔΔCT approach.
Transcription factor prediction
TRANSFAC is a database on transcription factors, as well as their genomic binding regions and DNA-binding profiling (http://transfac.gbf.de/TRANSFAC/).[30] Based on the database, transcription factors of RFX7 were inferred.
Data analysis
GraphPad Prism 9.0.1 (GraphPad Software, San Diego, CA) was used for statistical analyses and plotting the data. All values were presented as the means ± standard deviation. Student’s t-test or one-way analysis of variance was utilized to assess the significance of differences with the LSD post hoc test. Correlation analysis was achieved using a Pearson test. P < 0.01 indicated statistical significance. All experiments were repeated three times.
RESULTS
Aberrant transcriptome programs in children with persistent atopic asthma
In this study, transcriptome profiling of 36 children with persistent atopic asthma and 33 healthy children was performed. First, the characteristics of childhood asthma were investigated using the transcriptome program. The DEGs were selected in childhood asthma versus healthy controls. Consequently, 341 DEGs presented notable downregulation, with 361 presenting a notable upregulation in childhood asthma [Figure 1a and b]. In particular, the first 20 down-or upregulated DEGs are shown in Figure 1c and Table 1. Subsequently, the biological roles of the DEGs were evaluated. The biological processes underlying asthma include ECM organization, epidermal development, secretion by cells, response to wounding, peptidase activity, and fibroblast growth factor receptor signaling pathway [Figure 1d]. Cellular components such as extracellular vesicles or exosomes, integral components of membrane, plasma membrane, collagen-containing ECM, secretory granules, or vesicles may be modulated by DEGs [Figure 1e]. In addition, such components may possess key molecular functions such as protease binding, anion transmembrane transporter activity, and symporter activity [Figure 1f]. The crucial KEGG pathways, for example, oxidative phosphorylation, thermogenesis, ECM–receptor interaction, bile secretion, and metabolic pathways, were remarkably enriched by the DEGs [Figure 1g]. These findings unveil the key functions of the DEGs in childhood asthma.
| DEGs | Log2 fold change | P-value | q-value | Childhood asthma | Healthy controls |
|---|---|---|---|---|---|
| IGLL5 | −1.56119 | 0.01053 | 0.21222 | 7.64197 | 9.20316 |
| LOC96610 | −1.12679 | 0.00522 | 0.1673 | 5.30906 | 6.43585 |
| IGLL1 | −1.12555 | 0.00652 | 0.17827 | 5.6717 | 6.79725 |
| DEFB4A | −1.03844 | 0.00921 | 0.20256 | 9.78948 | 10.8279 |
| LOC100653210 | −1.0197 | 0.01767 | 0.261 | 5.36642 | 6.38612 |
| HLA-DOA | −0.95908 | 0.0000569 | 0.02134 | 8.62457 | 9.58365 |
| C4orf7 | −0.91864 | 0.03992 | 0.34828 | 6.35734 | 7.27598 |
| CSF3 | −0.8853 | 0.00946 | 0.20358 | 6.23205 | 7.11735 |
| FCGR3A | −0.88357 | 0.01002 | 0.20814 | 7.38137 | 8.26494 |
| HLA-DQA1 | −0.87987 | 0.00438 | 0.15556 | 9.42184 | 10.3017 |
| TMIGD2 | −0.871 | 0.00000143 | 0.00223 | 5.93525 | 6.80625 |
| KLRB1 | −0.86044 | 0.0000236 | 0.01815 | 6.88996 | 7.75041 |
| TCL1A | −0.85326 | 0.10775 | 0.49834 | 5.60882 | 6.46208 |
| CXCL13 | −0.82792 | 0.01044 | 0.21173 | 4.94867 | 5.77659 |
| CXCR6 | −0.82717 | 0.00000161 | 0.00223 | 6.28029 | 7.10746 |
| CCL5 | −0.82554 | 0.0000837 | 0.02536 | 7.5611 | 8.38664 |
| C1QB | −0.77734 | 0.00057 | 0.06121 | 8.70762 | 9.48496 |
| TNF | −0.76944 | 0.00057 | 0.06121 | 5.93691 | 6.70636 |
| FCRL5 | −0.76328 | 0.00667 | 0.17998 | 4.9119 | 5.67517 |
| VNN2 | −0.76095 | 0.00068 | 0.06952 | 8.58503 | 9.34598 |
| CST1 | 3.414466 | 0.0000000158 | 0.00011 | 7.74491 | 4.33045 |
| ITLN1 | 2.250443 | 0.0000154 | 0.01526 | 7.30274 | 5.0523 |
| CPA3 | 1.639058 | 0.000000353 | 0.00098 | 6.64493 | 5.00587 |
| TPSD1 | 1.378537 | 0.00000607 | 0.00765 | 6.49696 | 5.11843 |
| TPSAB1 | 1.376886 | 0.00000983 | 0.01136 | 6.53181 | 5.15493 |
| CTSG | 1.254341 | 0.000000127 | 0.00044 | 5.6366 | 4.38226 |
| CCL26 | 1.227964 | 0.00036 | 0.05253 | 6.26938 | 5.04141 |
| CCK | 1.152776 | 0.0000000117 | 0.00011 | 6.61707 | 5.4643 |
| CAPN14 | 1.082101 | 0.00016 | 0.03526 | 6.21819 | 5.13609 |
| KRT6A | 1.058431 | 0.01829 | 0.26374 | 9.91591 | 8.85748 |
| NTRK2 | 1.040872 | 0.00000157 | 0.00223 | 5.68855 | 4.64768 |
| CDH26 | 1.025739 | 0.0000312 | 0.01815 | 9.76298 | 8.73724 |
| ALOX15 | 1.017638 | 0.0000122 | 0.01301 | 9.13652 | 8.11888 |
| PRB4 | 0.96787 | 0.02991 | 0.31655 | 6.24727 | 5.2794 |
| CEBPE | 0.961805 | 0.00053 | 0.06029 | 5.96152 | 4.99971 |
| SCARNA5 | 0.915634 | 0.00012 | 0.03073 | 7.16332 | 6.24769 |
| FAM25A | 0.883917 | 0.02119 | 0.28122 | 6.76177 | 5.87786 |
| CYP1B1 | 0.866894 | 0.0106 | 0.21295 | 7.39275 | 6.52585 |
| PRB1 | 0.85142 | 0.03917 | 0.34747 | 6.46638 | 5.61496 |
| CD36 | 0.846708 | 0.0005 | 0.05982 | 8.2292 | 7.3825 |
Machine learning models based on RFX family members for diagnosing childhood asthma. DEGs: Differentially expressed genes, RFX: Regulatory factor X, IGLL5: Immunoglobulin lambda-like polypeptide 5, LOC96610: Hypothetical gene, IGLL1: Immunoglobulin lambda-like polypeptide 1, DEFB4A: Defensin beta 4A, HLA-DOA: Major histocompatibility complex, class II, DO alpha, C4orf7: Follicular dendritic cell-secreted protein, CSF3: Colony-stimulating factor 3, FCGR3A: Fc gamma receptor IIIa, HLA-DQA1: Major histocompatibility complex, class II, DQ alpha 1, TMIGD2: Transmembrane and immunoglobulin domain containing 2, KLRB1: Killer cell lectin-like receptor B1, TCL1A: TCL1 family AKT coactivator A, CXCL13: C-X-C motif chemokine ligand 13, CXCR6: C-X-C motif chemokine receptor 6, CCL5: C-C motif chemokine ligand 5, C1QB: Complement C1q B chain, TNF: Tumor necrosis factor, FCRL5: Fc receptor-like 5, VNN2: Vanin 2, CST1: Cystatin SN, ITLN1: Intelectin 1, CPA3: Carboxypeptidase A3, TPSD1: Tryptase delta 1, TPSAB1: Tryptase alpha/beta 1, CTSG: Cathepsin G, CCL26: C-C motif chemokine ligand 26, CCK: Cholecystokinin, CAPN14: Calpain 14, KRT6A: Keratin 6A, NTRK2: Neurotrophic receptor tyrosine kinase 2, CDH26: Cadherin 26, ALOX15: Arachidonate 15-lipoxygenase, PRB4: Proline-rich protein BstNI subfamily 4, CEBPE: CCAAT enhancer-binding protein epsilon, SCARNA5: Small Cajal body-specific RNA 5, FAM25A: Family with sequence similarity 25 member A, CYP1B1: Cytochrome P450 family 1 subfamily B member 1, PRB1: Proline-rich protein BstNI subfamily 1, CD36: Cluster of differentiation 36
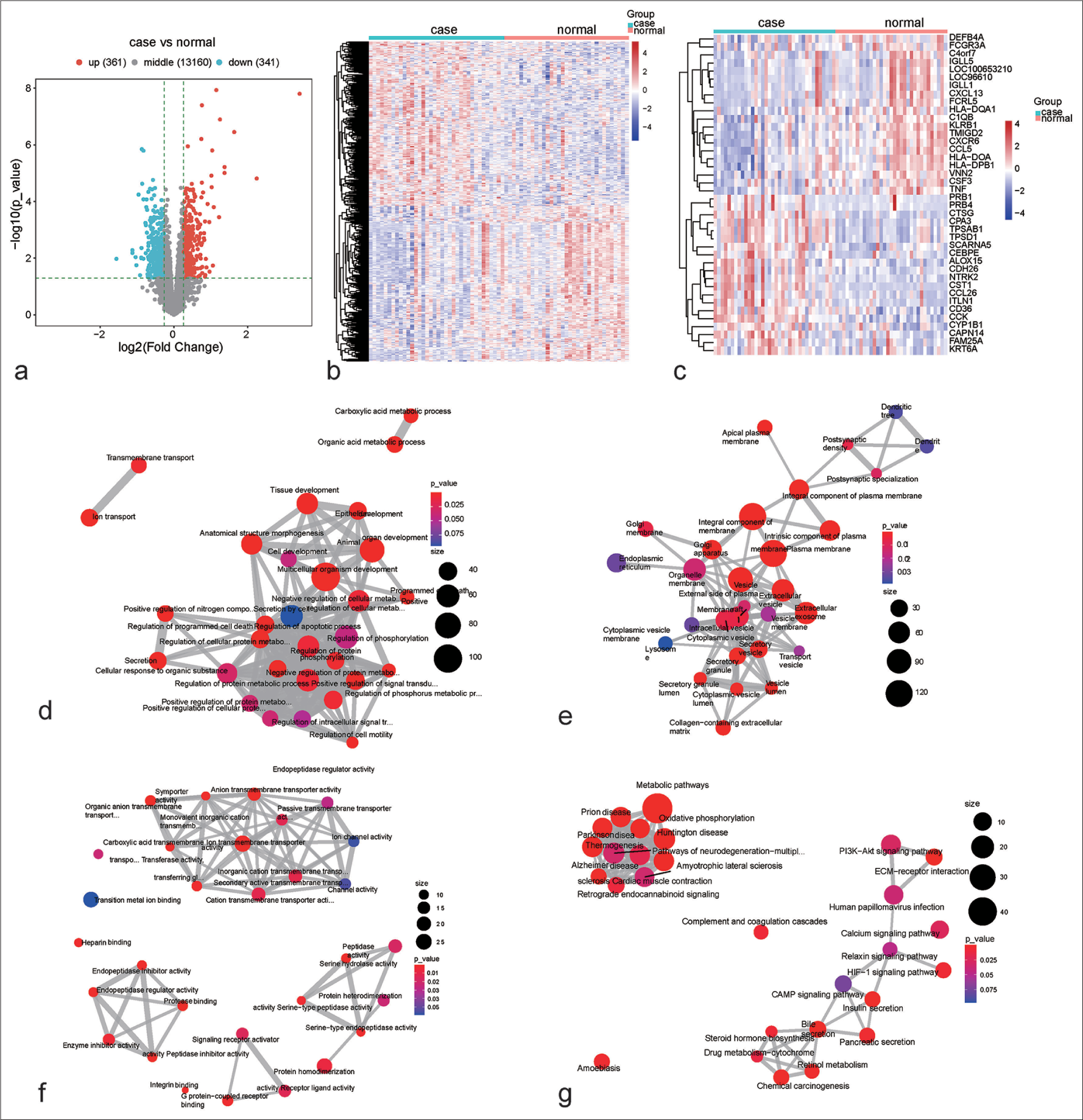
- Aberrant transcriptome programs in children with persistent atopic asthma. (a) Selection of genes with differential expression in children with persistent atopic asthma versus healthy controls. Blue, downregulation; red, upregulation; and gray, no significance. (b) Transcriptome patterns of the identified differentially expressed genes (DEGs) in persistent atopic asthma and healthy controls. Blue to red denotes down-to upregulated expression. (c) Transcriptome patterns of the first 20 down-to upregulated genes in persistent atopic asthma in comparison with healthy controls. (d-f) The biological process, cellular component, or molecular function network based on the DEGs. From blue to red means that P-value goes down. The larger the bubble, the more genes are enriched. (g) The kyoto encyclopedia of genes and genomes pathway network based on the DEGs. Machine learning models based on RFX family members for diagnosing childhood asthma. DEGs: Differentially expressed genes, RFX: Regulatory factor X, IGLL5: Immunoglobulin lambda-like polypeptide 5, LOC96610: Hypothetical gene, IGLL1: Immunoglobulin lambda-like polypeptide 1, DEFB4A: Defensin beta 4A, HLA-DOA: Major histocompatibility complex, class II, DO alpha, C4orf7: Follicular dendritic cell-secreted protein, CSF3: Colony-stimulating factor 3, FCGR3A: Fc gamma receptor IIIa, HLADQA1: Major histocompatibility complex, class II, DQ alpha 1, TMIGD2: Transmembrane and immunoglobulin domain containing 2, KLRB1: Killer cell lectin-like receptor B1, TCL1A: TCL1 family AKT coactivator A, CXCL13: C-X-C motif chemokine ligand 13, CXCR6: C-X-C motif chemokine receptor 6, CCL5: C-C motif chemokine ligand 5, C1QB: Complement C1q B chain, TNF: Tumor necrosis factor, FCRL5: Fc receptor-like 5, VNN2: Vanin 2, CST1: Cystatin SN, ITLN1: Intelectin 1, CPA3: Carboxypeptidase A3, TPSD1: Tryptase delta 1, TPSAB1: Tryptase alpha/beta 1, CTSG: Cathepsin G, CCL26: C-C motif chemokine ligand 26, CCK: Cholecystokinin, CAPN14: Calpain 14, KRT6A: Keratin 6A, NTRK2: Neurotrophic receptor tyrosine kinase 2, CDH26: Cadherin 26, ALOX15: Arachidonate 15-lipoxygenase, PRB4: Proline-rich protein BstNI subfamily 4, CEBPE: CCAAT enhancer-binding protein epsilon, SCARNA5: Small Cajal body-specific RNA 5, FAM25A: Family with sequence similarity 25 member A, CYP1B1: Cytochrome P450 family 1 subfamily B member 1, PRB1: Proline-rich protein BstNI subfamily 1, CD36: Cluster of differentiation 36 , HLA-DPB1:major histocompatibility complex, class II, DP beta 1
Machine learning models based on RFX family members for diagnosing childhood asthma
Among RFX family members, RFX3, RFX7, and RFX-associated protein (RFXAP) presented differential expression in childhood asthma. On the basis of RFX3, RFX7, and RFXAP, the RFX ssGSEA score in childhood asthma and control specimens was estimated. The RFX ssGSEA score was remarkably higher in childhood asthma than in controls [Figure 2a and b], indicating the activation of RFX in childhood asthma. Thus, RFX family members can be used to diagnose childhood asthma. In addition, SVM, logistic regression, and random forest-based machine-learning models were established. Subsequently, the diagnostic efficacy was evaluated. In the discovery set, the AUC values of the SVM, logistic regression, and random forest-based models were 0.83, 0.83, and 1.00, respectively [Figure 2c-e]. Reproducibility was further proven in the verification set. Consequently, the AUC values of the SVM, logistic regression and random forest-based models were 0.58, 0.64, and 0.67, respectively. Overall, the random forest-based model presented the best diagnostic efficiency in childhood asthma. The efficiency of single RFX family members in childhood asthma diagnosis was also assessed. The AUC values of RFX3, RFX7, and RFXAP were 0.657, 0.724, and 0.766, respectively, in the discovery set, and 0.617, 0.775, and 0.566, respectively, in the verification set [Figure 2f-h]. Only the larger area under the curve of the G-chart indicates that RFX7 has the most favorable diagnostic effect on childhood asthma. Hence, among the RFX family members, RFX7 had the most favorable efficacy in diagnosing childhood asthma.
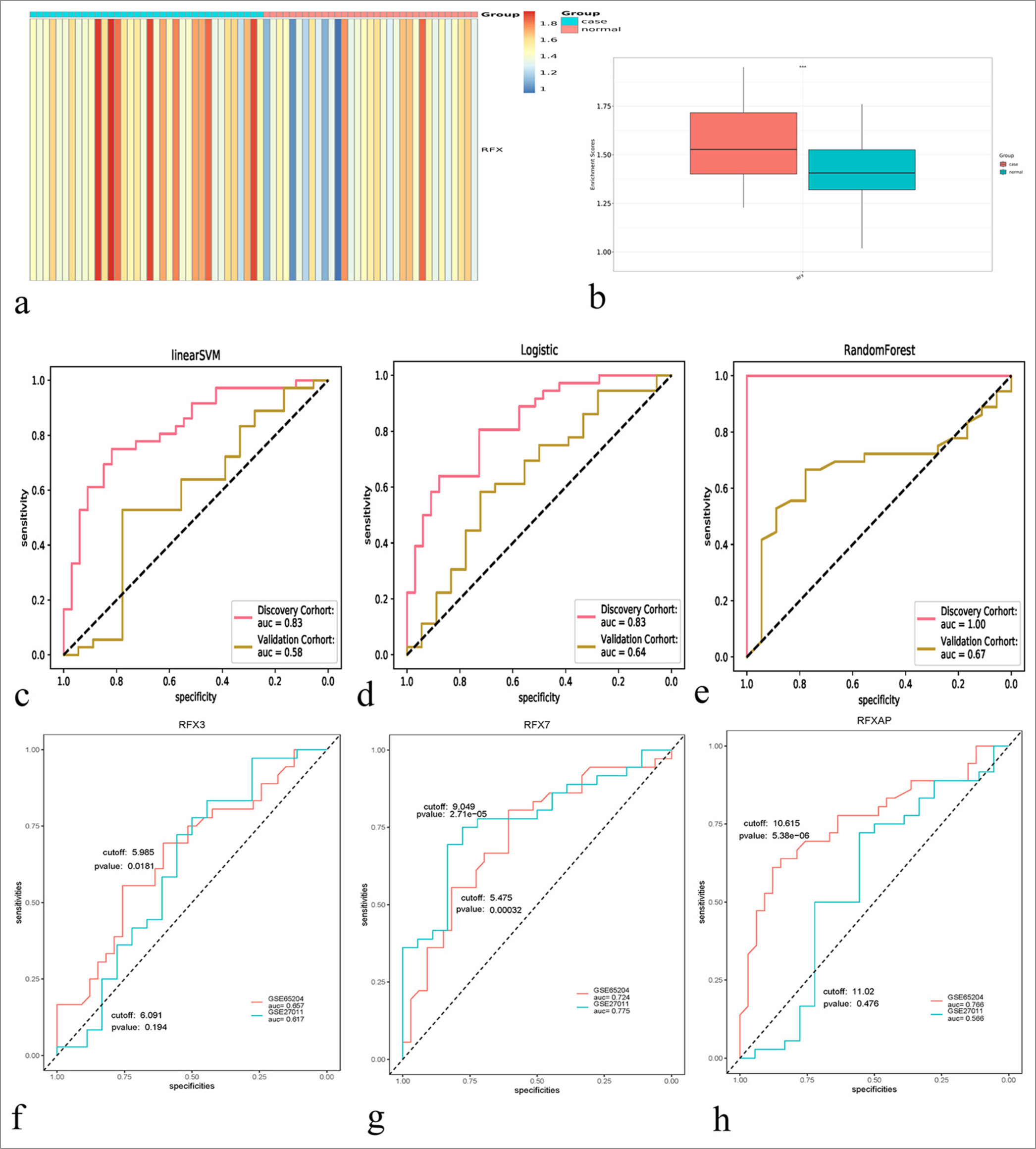
- Machine learning models based on the regulatory factor X (RFX) family members for diagnosing childhood asthma. (a) The single– sample gene set enrichment approach (ssGSEA) score of the RFX family members in childhood asthma and healthy controls. Blue to red represents low-to-high ssGSEA score. (b) Comparison of the RFX ssGSEA score in childhood asthma versus healthy controls. ✶✶✶P < 0.001. (c-e) Receiver operating characteristics for the evaluation of the diagnostic efficacy of linear support vector machine, logistic regression, and random forest–based machine learning models based on the RFX family members in the discovery and verification sets. (f-h) Assessment of the performance of RFX3, RFX7, and RFX-associated protein in diagnosing childhood asthma in the two sets.
RFX7 presents remarkable overexpression in asthma cell models
A total of 50 ng/mL PDGF-BB-induced ASMCs were utilized for the establishment of asthma cell models. RFX7 expression was greatly activated in PDGF-BB-induced ASMCs versus control [Figure 3a and b]. For further evaluating the role of RFX7 in asthma, three specific siRNAs targeting RFX7 were transfected into ASMCs. RFX7 was efficiently knocked out in ASMCs by its siRNAs compared with si-NC [Figure 3c-e]. Considering that RFX7 siRNA#3 had the best knockout effect, it was used for follow-up experiments.
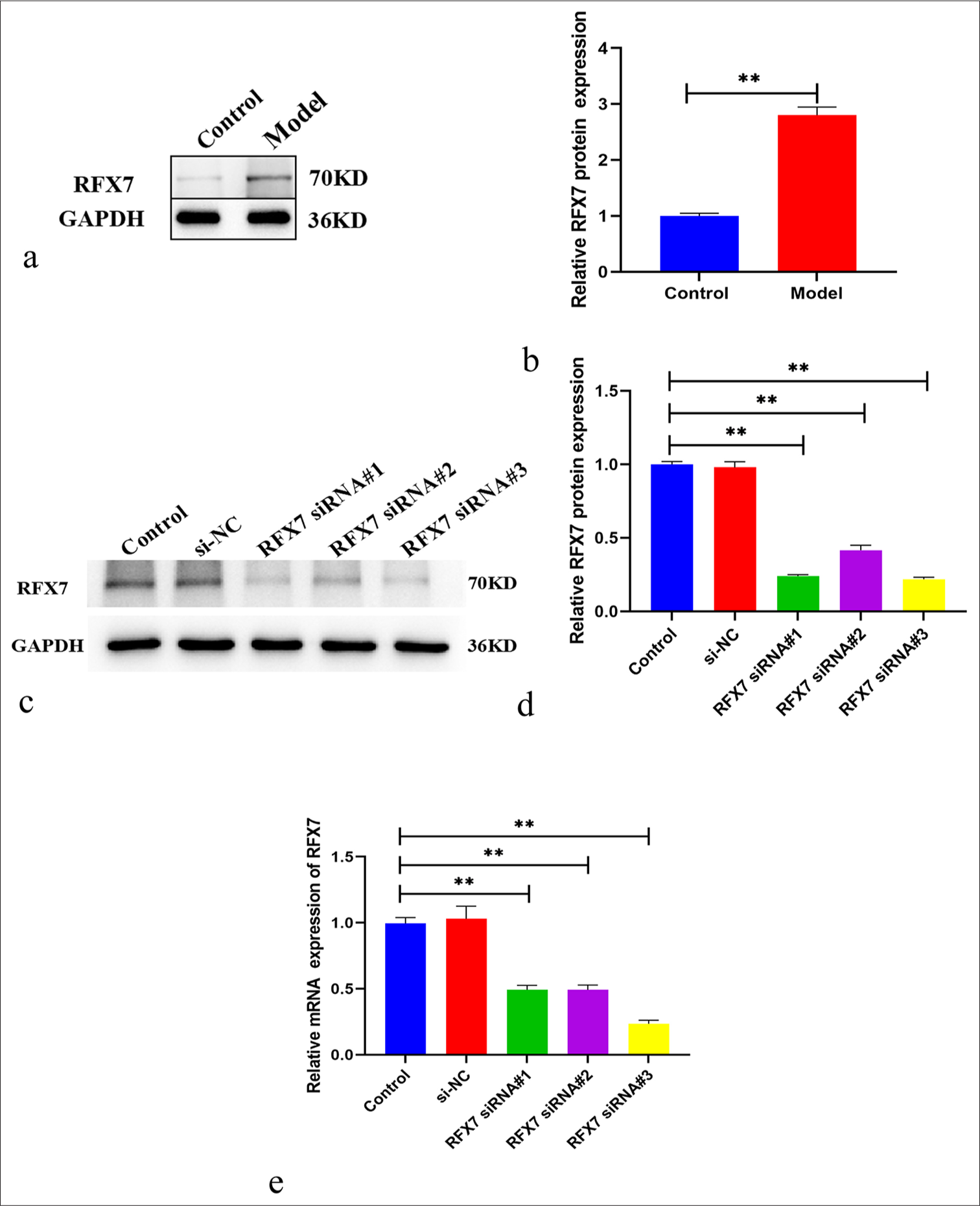
- Regulatory factor X (RFX7) displays the notable overexpression in asthma cell models. (a) Representative Western blotting images of RFX7 in airway smooth muscle cells (ASMCs) administrated with 50 ng/mL platelet-derived growth factor-BB (PDGF-BB) or control. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as a reference control. (b) Quantification analysis of RFX7 expression in control or PDGF-BB-induced ASMCs. (c) Representative Western blotting images of RFX7 in ASMCs in the context of small interfering-negative control (si- NC), RFX7 Small interfering RNAs (siRNA)#1, RFX7 siRNA#2, or RFX7 siRNA#3 transfection. (d) Quantification analysis of RFX7 expression in si-NC, RFX7 siRNA#1-, RFX7 siRNA#2-, or RFX7 siRNA#3-transfected ASMCs. (e) Representative reverse transcription quantitative polymerase chain reaction of RFX7 in ASMCs in the context of si-NC, RFX7 siRNA#1, RFX7 siRNA#2, or RFX7 siRNA#3 transfection. ✶✶ compared with the control group at P < 0.01. The experiment was repeated 3 times, n = 3.
Suppression of RFX7 alleviates PDGF-BB-induced ASMC proliferation
The improved proliferative capacity of ASMCs is greatly linked with asthma.[12] As expected, ASMC proliferation was prominently heightened in the context of PDGF-BB stimulation [Figure 4a and b]. In addition, we focused on whether RFX7 influenced PDGF-BB-stimulated ASMC proliferation. As a result, RFX7 knockdown efficiently alleviated the proliferative ability of ASMCs in the context of PDGF-BB.
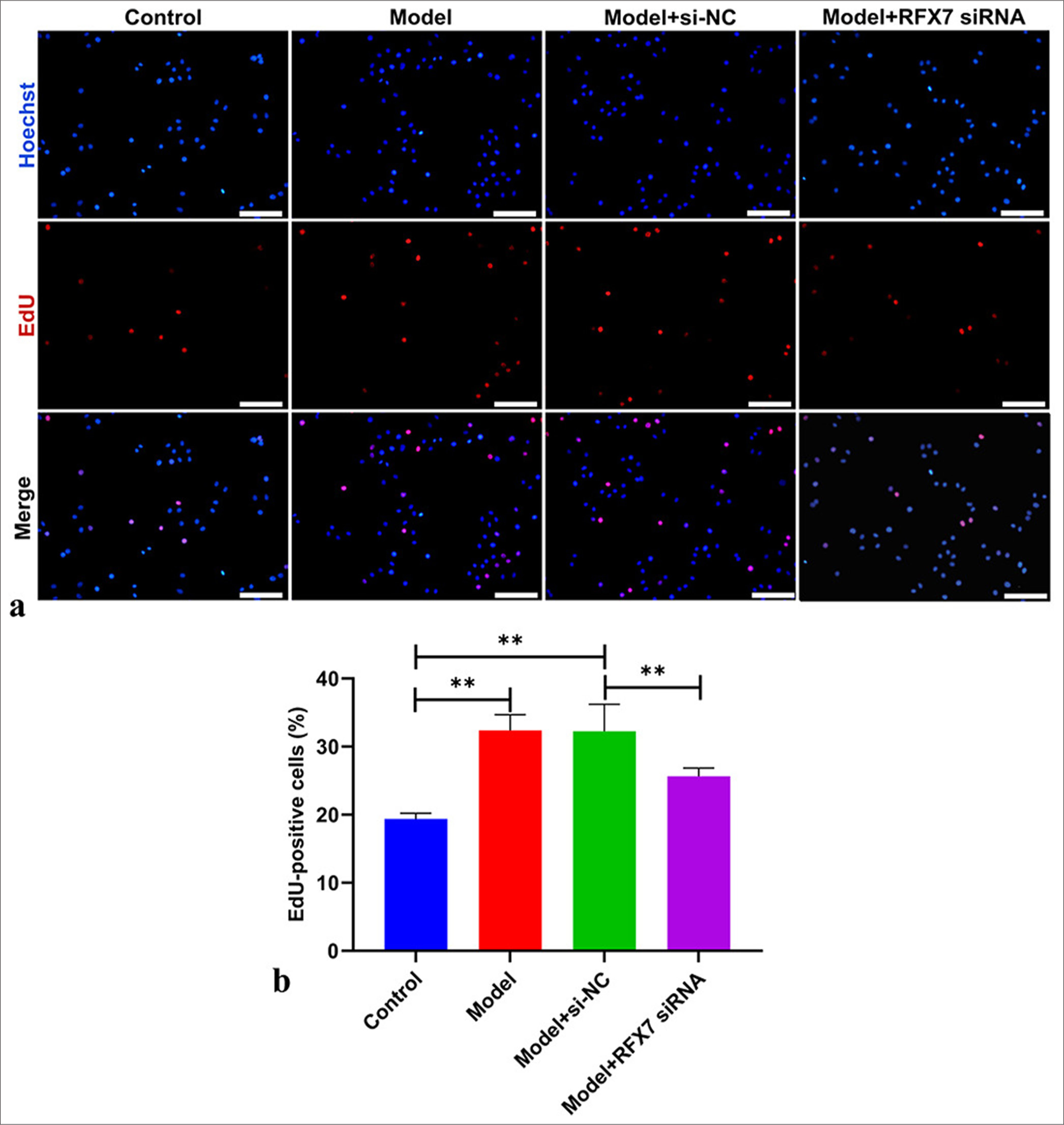
- Regulatory factor X7 (RFX7) knockdown alleviates platelet-derived growth factor-BB (PDGF-BB)-triggered airway smooth muscle cell (ASMC) proliferation. (a) Representative images of 5-ethyny-2'-deoxyuridine (EdU) assay in ASMCs in the context of PDGF-BB administration or RFX7 Small interfering RNAs (siRNA) transfection. Scale bar, 50 μm. (b) Quantification analysis of EdU-positive ASMCs with PDGF-BB administration or RFX7 siRNA transfection. ✶✶P < 0.01. The experiment was repeated 3 times, n = 3.
RFX7 knockdown ameliorates ASMC migration stimulated by PDGF-BB
The excessive migration of ASMCs directly results in airway remodeling in asthma.[31] The migrative capacity of ASMCs was evaluated using wound healing and Transwell assays. Consequently, closure ratios were greatly improved by PDGF-BB stimulation in ASMCs [Figure 5a and b]. RFX7 inhibition efficiently attenuated the closure ratios of ASMCs under PDGF-BB administration. In addition, the number of migrative ASMCs was prominently elevated following PDGF-BB administration [Figure 5c and d]. RFX7 suppression greatly reduced the number of migrative ASMCs stimulated by PDGF-BB. Hence, silencing RFX7 impaired ASMC migration under PDGF-BB stimulation.
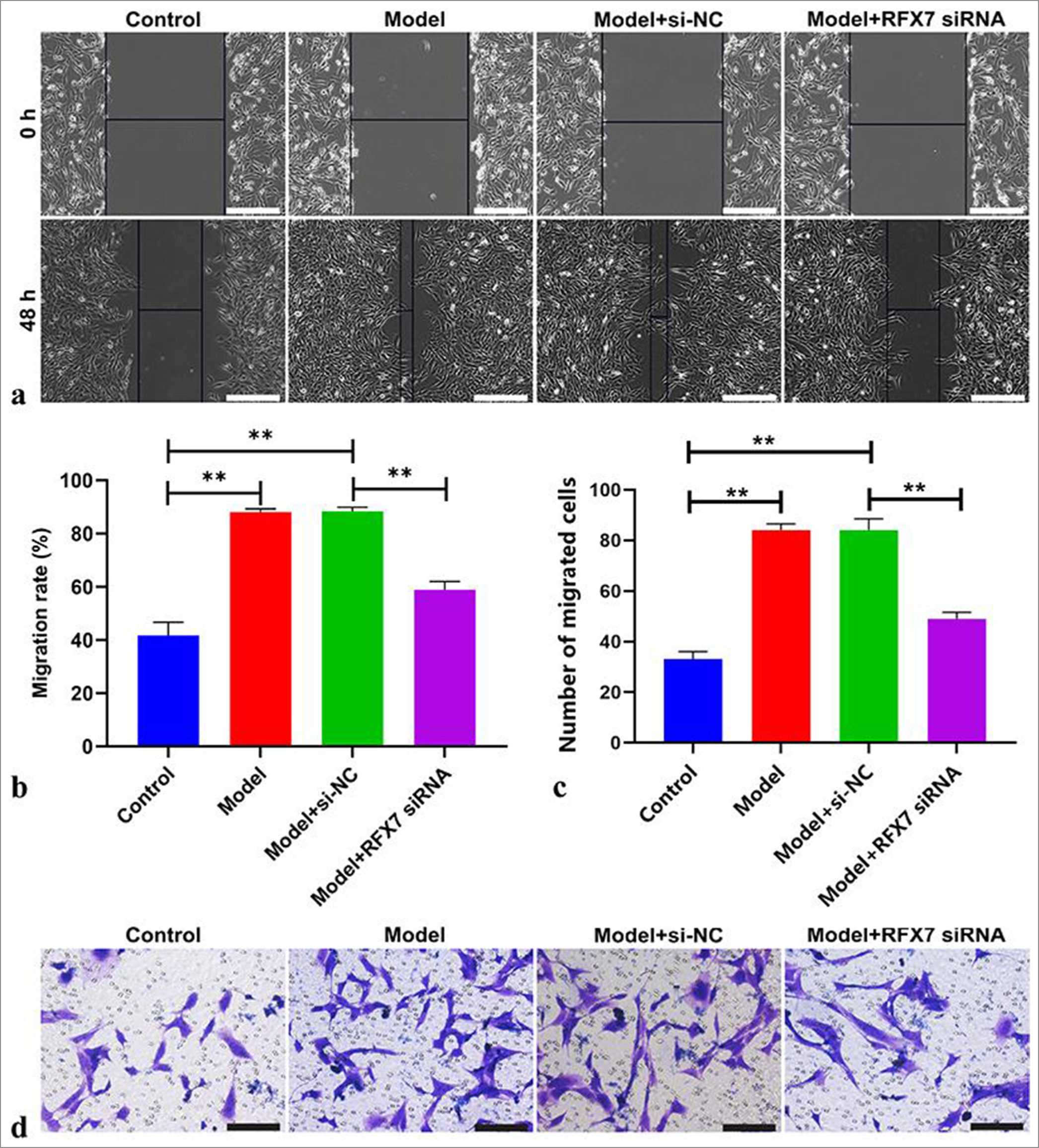
- Targeting regulatory factor X7 (RFX7) ameliorates airway smooth muscle cell (ASMC) migration stimulated by platelet-derived growth factor-BB (PDGF-BB). (a) Representative photographs of wound healing assay in ASMCs with PDGF-BB stimulation or RFX7 small interfering RNAs (siRNA) transfection. Scale bar, 100 μm. (b) Quantification of the closure ratios of ASMCs with regard to PDGFBB treatment or RFX7 siRNA transfection. (c) Representative photographs of Transwell assay in ASMCs stimulated with PDGF-BB or transfected with RFX7 siRNAs. Cell mobility = (initial scratch width − final scratch width)/initial scratch width × 100%. (d) Quantification of migrative ASMCs with regard to PDGF-BB administration or transfection with RFX7 siRNAs. Scale bar, 50 μm.; ✶✶P < 0.01. The experiment was repeated 3 times, n = 3.
Silencing RFX7 weakens PDGF-BB-stimulated ASMC remodeling
An in-depth assessment was conducted to determine the influence of RFX7 on ASMC remodeling. α-SMA [Figure 6a and b] and N-cadherin [Figure 6c and d] levels displayed a remarkable increase in ASMCs with regard to PDGF-BB stimulation. Meanwhile, E-cadherin levels presented a reduction in PDGF-BB-stimulated ASMCs [Figure 6e and f]. RFX7 suppression efficiently decreased α-SMA and N-cadherin levels as well as increased E-cadherin levels in PDGF-BB-induced ASMCs. The abovementioned data indicated that silencing RFX7 alleviated PDGF-BB-stimulated ASMC remodeling.
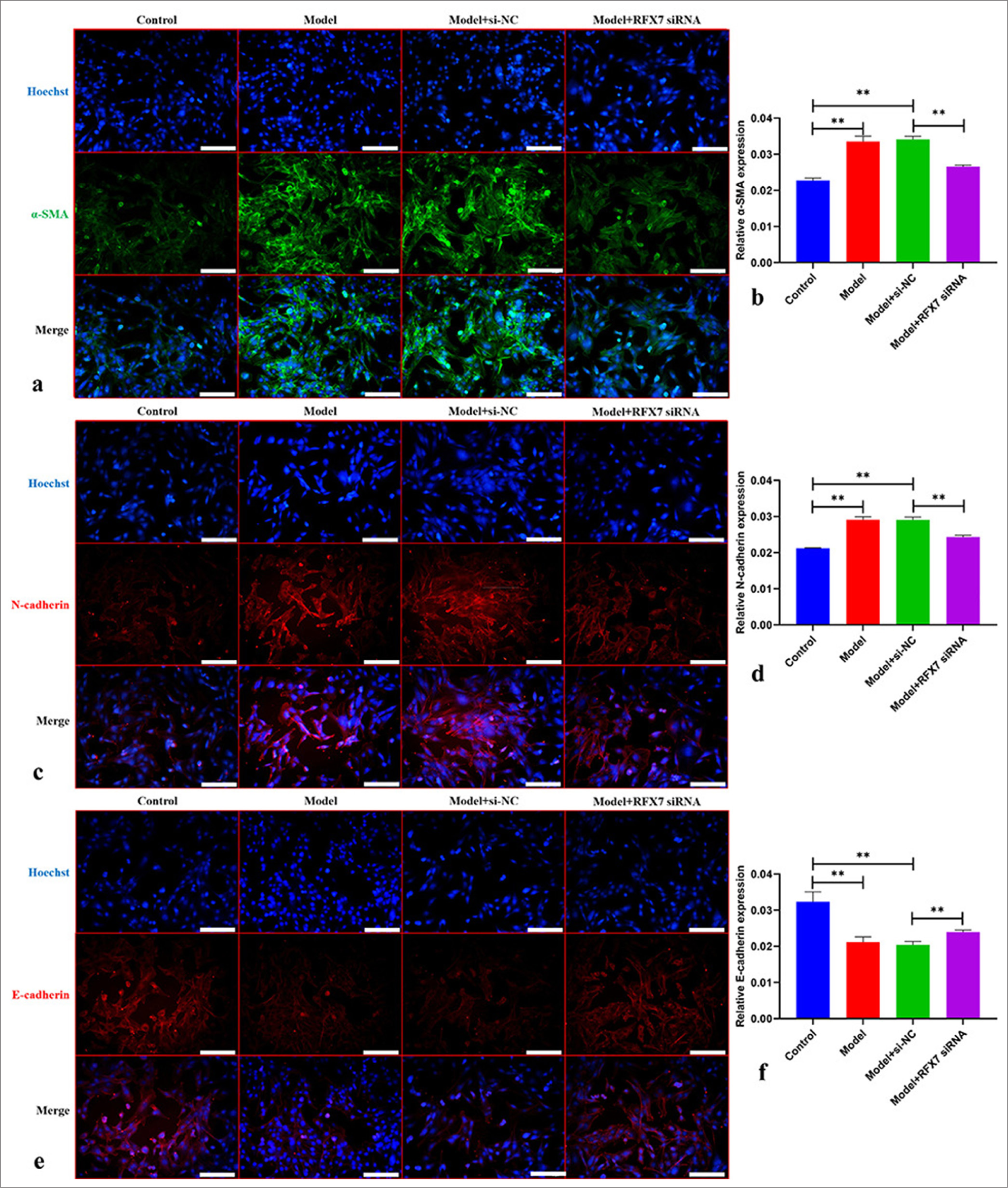
- Silencing regulatory factor X7 (RFX7) weakens platelet-derived growth factor-BB (PDGF-BB)-stimulated airway smooth muscle cell (ASMC) remodeling. (a) Representative photographs of immunofluorescence on alpha smooth muscle actin (α-SMA) in ASMCs with PDGFBB stimulation or RFX7 small interfering RNAs (siRNA) transfection. Scale bar, 150 μm. (b) Quantification analysis on α-SMA in ASMCs with PDGF-BB stimulation or RFX7 siRNA transfection. (c) Representative photographs of immunofluorescence on N-cadherin in ASMCs administrated with PDGF-BB or transfected with RFX7 siRNAs. Scale bar, 150 μm. (d) Quantification analysis on N-cadherin in ASMCs administrated with PDGF-BB or transfected with RFX7 siRNAs. (e) Representative photographs of immunofluorescence on E-cadherin in ASMCs with regard to PDGF-BB stimulation or RFX7 siRNA transfection. Scale bar, 50 μm. (f) Quantification analysis on E-cadherin in ASMCs with regard to PDGF-BB stimulation or RFX7 siRNA transfection. ✶✶P < 0.01. The experiment was repeated 3 times, n = 3.
Targeting RFX7 decreases inflammatory response in PDGF-BB-stimulated ASMCs
RFX ssGSEA score and RFX7 positively correlated with neutrophil infiltration in childhood asthma [Figure 7a]. Pro-inflammatory cytokines: TNF-α, IL-1 β, and IL-6 presented notably elevated levels in ASMCs with regard to PDGF-BB administration [Figure 7b-d]. RFX7 suppression efficiently decreased the levels of TNF-α, IL-1 β, and IL-6 in ASMCs stimulated by PDGF-BB. Collectively, RFX7 silencing decreased inflammatory response in PDGF-BB-induced ASMCs
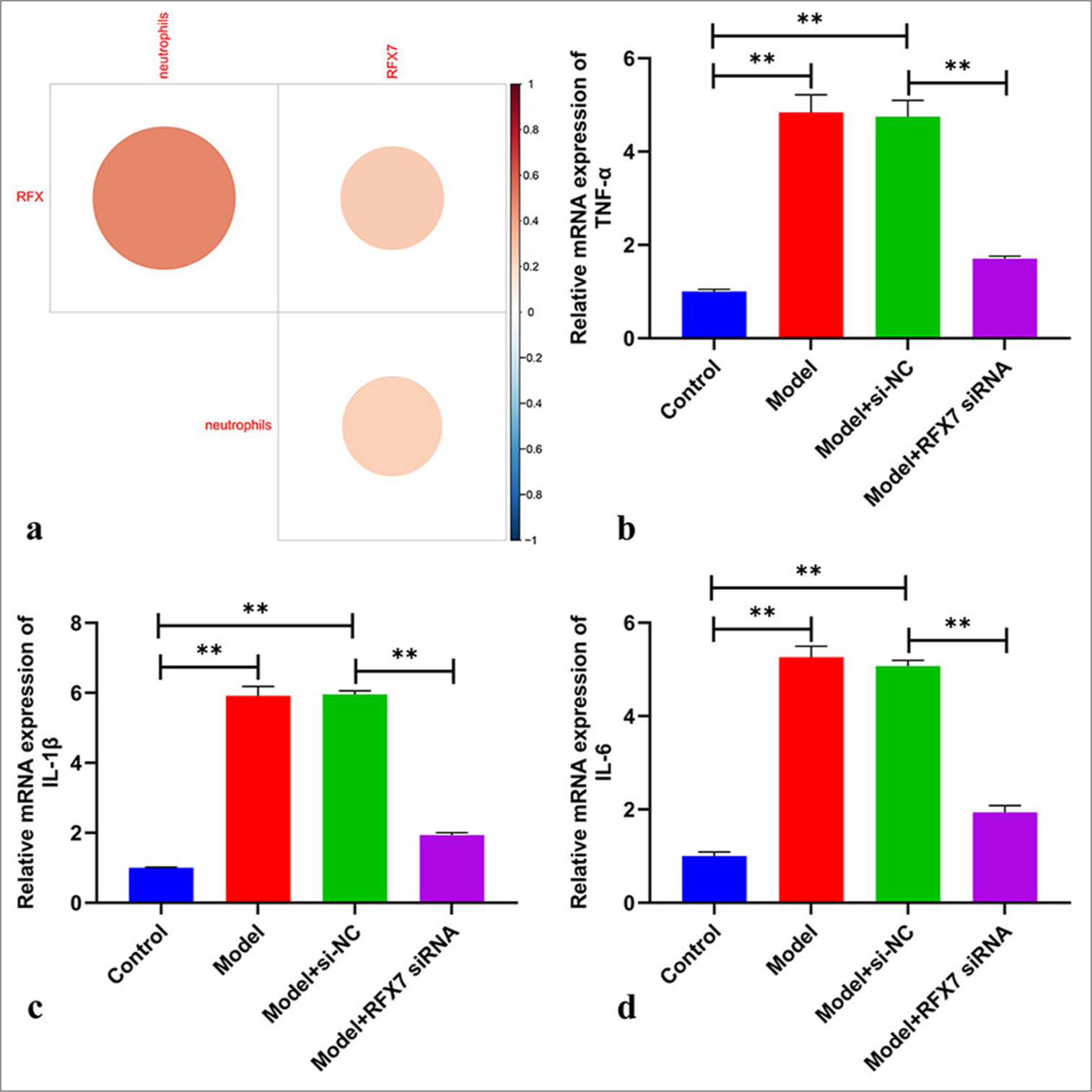
- Targeting regulatory factor X7 (RFX7) decreases inflammatory response in platelet-derived growth factor-BB (PDGF-BB)-stimulated ASMCs. (a) Correlation analysis on the RFX single–sample gene set enrichment approach score, RFX7, and neutrophils in childhood asthma. (b-d) reverse transcription quantitative polymerase chain reaction on the detection of mRNA levels of tumor necrosis factor alpha-like, interleukin 1 beta, and interleukin 6in ASMCs with PDGF-BB stimulation or transfection with RFX7 siRNAs. ✶✶P < 0.01. The experiment was repeated 3 times, n = 3.
Prediction of the transcription factors of RFX7
A total of 59 transcription factors of RFX7 were estimated, including AT-rich interaction domain 3A, bromodomain containing 2, bromodomain containing 4, cyclin dependent kinase 7, cyclin-dependent kinase 9, CCAAT enhancer-binding protein beta, cAMP responsive element-binding protein 1, CREB-binding protein, CCCTC-binding factor, CCCTC-binding factor like, E2F transcription factor 6, E74 like ETS transcription factor 1, ETS proto-oncogene 1, transcription factor, Fli-1 proto-oncogene, ETS transcription factor, Forkhead box A2, Forkhead box P1, GATA-binding protein 1, host cell factor C1, histone deacetylase 2, Hes-related family bHLH transcription factor with YRPW motif 1, hepatocyte nuclear factor 4 alpha, hepatocyte nuclear factor 4 gamma, lysine acetyltransferase 2B, lysine demethylase 5B, KLF transcription factor 9, lamin B1, MYC-associated zinc finger protein, mediator complex subunit 12, MYB proto-oncogene, transcription factor, myogenic determining factor 3, nuclear factor I C, Notch receptor 1, Paired box 5, PHD finger protein 8, RNA polymerase II subunit A, POU class 2 homeobox 1, RAD21 cohesin complex component, RB-binding protein 5, histone lysine methyltransferase complex subunit, RUNX family transcription factor 3, Retinoid X receptor gamma, SET domain bifurcated histone lysine methyltransferase 1, SIN3 transcription regulator family member A, structural maintenance of chromosomes 1A, structural maintenance of chromosomes 3, Sp4 transcription factor, Spi-1 proto-oncogene, SRC protooncogene, non-receptor tyrosine kinase, serum response factor, STAG1 cohesin complex component, signal transducer and activator of transcription 1, TATA-box-binding protein-associated factor 1, TATA-box-binding protein, transcription factor 3, transcription factor AP-4, tumor protein p63, upstream binding transcription factor, WD repeat domain 5, YY1 transcription factor, and zinc finger protein 263 [Table 2]. RFX7 might be transcriptionally modulated by the above-mentioned transcription factors.
| Transcription factors of RFX7 | |
|---|---|
| Transcription factors | ARID3A, BRD2, BRD4, CDK7, CDK9, CEBPB, CREB1, CREBBP, CTCF, CTCFL, E2F6, ELF1, ETS1, FLI1, FOXA2, FOXP1, GATA1, HCFC1, HDAC2, HEY1, HNF4A, HNF4G, KAT2B, KDM5B, KLF9, LMNB1, MAZ, MED12, MYB, MYOD1, NFIC, NOTCH1, PAX5, PHF8, POLR2A, POU2F1, RAD21, RBBP5, RUNX3, RXRG, SETDB1, SIN3A, SMC1A, SMC3, SP4, SPI1, SRC, SRF, STAG1, STAT1, TAF1, TBP, TCF3, TFAP4, TP63, UBTF, WDR5, YY1, and ZNF263 |
RFX7: Regulatory factor X7, ARID3A: AT-rich interaction domain 3A, BRD2: Bromodomain containing 2, BRD4: Bromodomain containing 4, CDK7: Cyclin-dependent kinase 7, CDK9: Cyclin-dependent kinase 9, CEBPB: CCAAT enhancer-binding protein beta, CREB1: cAMP responsive element-binding protein 1, CREBBP: CREB-binding protein, CTCF: CCCTC-binding factor, CTCFL: CCCTC-binding factor like, E2F6: E2F transcription factor 6, ELF1: E74 like ETS transcription factor 1, ETS1: ETS proto-oncogene 1, transcription factor, FLI1: Fli-1 proto-oncogene, ETS transcription factor, FOXA2: Forkhead box A2, FOXP1: Forkhead box P1, GATA1: GATA-binding protein 1, HCFC1: Host cell factor C1, HDAC2: Histone deacetylase 2, HEY1: Hes-related family bHLH transcription factor with YRPW motif 1, HNF4A: Hepatocyte nuclear factor 4 alpha, HNF4G: Hepatocyte nuclear factor 4 gamma, KAT2B: Lysine acetyltransferase 2B, KDM5B: Lysine demethylase 5B, KLF9: KLF transcription factor 9, LMNB1: Lamin B1, MAZ: MYC-associated zinc finger protein, MED12: Mediator complex subunit 12, MYB: MYB proto-oncogene, transcription factor, MYOD1: Myogenic determining factor 3, NFIC: Nuclear factor I C, NOTCH1: Notch receptor 1, PAX5: Paired box 5, PHF8: PHD finger protein 8, POLR2A: RNA polymerase II subunit A, POU2F1: POU class 2 homeobox 1, RAD21: RAD21 cohesin complex component, RBBP5: RB-binding protein 5, histone lysine methyltransferase complex subunit, RUNX3: RUNX family transcription factor 3, RXRG: Retinoid X receptor gamma, SETDB1: SET domain bifurcated histone lysine methyltransferase 1, SIN3A: SIN3 transcription regulator family member A, SMC1A: Structural maintenance of chromosomes 1A, SMC3: Structural maintenance of chromosomes 3, SP4: Sp4 transcription factor, SPI1: Spi-1 proto-oncogene, SRC: SRC proto-oncogene, non-receptor tyrosine kinase, SRF: Serum response factor, STAG1: STAG1 cohesin complex component, STAT1: Signal transducer and activator of transcription 1, TAF1: TATA-box-binding protein associated factor 1, TBP: TATA-box-binding protein, TCF3: Transcription factor 3, TFAP4: Transcription factor AP-4, TP63: Tumor protein p63, UBTF: Upstream binding transcription factor, WDR5: WD repeat domain 5, YY1: YY1 transcription factor, ZNF263: Zinc finger protein 263
DISCUSSION
The etiology of asthma is complex, covering complicated genetic and environmental factors.[32] Nonetheless, the mechanisms underlying the disease need definitive characterization. In this study, the transcriptome program features of childhood asthma were unveiled, and a machine learning RFX-based model was built for childhood asthma diagnosis. The random forest model presented the most excellent diagnostic efficiency, which might be applied in clinical practice. The favorable efficacy of RFX7 in diagnosing childhood asthma was also proven.
Airway smooth muscle response exerts a crucial significance in asthma. For example, the crosstalk of CD34+ fibrocytes with ASMCs (a dominating structural component of the airway) triggers IL-8 generation and activates the Akt (Serine/threonine kinase 1)/PRAS40 (Proline-rich AKT substrate)/mTOR (mechanistic target of rapamycin kinase) pathway in asthma.[33] During the pathologic development of asthma, the proliferation and migration of ASMCs can be elicited by distinct stimuli (such as PDGF-BB), thereby increasing ASM thickness and inducing airway wall remodeling. Hence, suppressing aberrant ASMC proliferation and migration is a potential therapy against asthma. PDGF-BB can be secreted by activated platelets and endothelial, epithelial, glial, and inflammatory cells, which has been proven as a dominating stimulus for promoting ASMC proliferation and hypertrophy as well as ASMC migration.[34] The expression level of RFX7 was greatly elevated in PDGF-BB-stimulated ASMCs. Silencing RFX7 attenuated the proliferative and migrative capacities of ASMCs with regard to PDGF-BB. In addition, RFX7 inhibition efficiently decreased α-SMA and N-cadherin levels as well as increased E-cadherin levels in PDGF-BB-induced ASMCs. Accordingly, therapeutic targeting of RFX7 might be a potential therapeutic regimen for childhood asthma.
Asthma symptoms are triggered by airway inflammation, inducing multiple processes, for example, mucus generation, airway wall remodeling, and bronchial hyperresponsiveness.[35] Airway neutrophil infiltration has been extensively proven to be associated with asthma severity.[36] Notably, neutrophil infiltration is associated with asthma that is unresponsive to corticosteroids, a mainstay of asthma therapy.[37] Nevertheless, effective regimens to alleviate neutrophilic airway inflammation in serious asthma remain elusive. Our data indicated that RFX7 presented a positive interaction with neutrophil infiltration in childhood asthma. The pathological characteristics of asthma include persistent airway inflammation, infiltration of inflammatory cells, and the release of pro-inflammatory cytokines and mediators.[38] TNF-α, IL-1 β, and IL-6 are potent inflammatory cytokines that result in airway inflammation, influencing therapeutic outcomes in asthma.[39,40] Silencing RFX7 decreased the levels of TNF-α, IL-1 β, and IL-6 in PDGF-BB-stimulated ASMCs, thereby alleviating inflammatory response during asthma.
Nonetheless, several limitations should be pointed out. In the follow-up study, the overexpression of RFX7 will be considered. Prospective cohorts are also necessary to further verify the diagnostic efficiency of the machine learning-based RFX model and RFX7 in childhood asthma. Furthermore, the transcriptional mechanisms and influence of RFX7 in the disease need in-depth investigations.
SUMMARY
Collectively, the random forest-based RFX model presented the best efficiency in diagnosing childhood asthma. Notably, RFX7 promoted the diagnosis of childhood asthma among RFX family members. RFX7 was proven to be upregulated in PDGF-BB-stimulated ASMCs; moreover, the suppression of RFX7 remarkably alleviated PDGF-BB-induced airway remodeling and inflammation. Hence, RFX7 was regarded as a therapeutic molecule for childhood asthma.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENT
Not applicable
ABBREVIATIONS
ANOVA: One-way analysis of variance
ASMCs: Airway smooth muscle cells
DEGs: Differentially expressed genes
ECM: Extracellular matrix
EdU: 5-Ethyny-2’-deoxyuridine
GO: Gene Ontology
GSEA: Gene set enrichment approach
GSVA: Gene set variation analysis
KEGG: Kyoto Encyclopedia of Genes and Genomes
MHCII: MHC class II
PDGF-BB: Platelet-derived growth factor BB
RFX: Regulatory factor X
ROCs: Receiver operating characteristic curves
RT-qPCR: Reverse transcription quantitative polymerase chain reaction
siRNAs: Small interfering RNAs
ssGSEA: Simple-sample GSEA
SVM: Support vector machine
AUTHOR CONTRIBUTIONS
YHW and FL: Designed the research; JWQ, JG and JTF: Performed the experiments; JM, TSD, and XLL: Analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by Ethics Committee of Ji’an Hospital, Shanghai East Hospital ([2021] Research Preliminary Review No. [04]), and all experiments complied with approved protocols.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: This project was supported by 2021 Jiangxi Provincial Department of Science and Technology Applied Research Cultivation Plan Project (No.20212BAG70005, Yahui Wu) and 2021 Science and Technology Special Project and Social Development Project in Ji’an City, Jiangxi Province (No.20211-025242,Yahui Wu).
References
- Residential greenspace and childhood asthma: An intra-city study. Sci Total Environ. 2023;857:159792.
- [CrossRef] [PubMed] [Google Scholar]
- Creatine kinase is decreased in childhood asthma. Am J Respir Crit Care Med. 2023;207:544-52.
- [CrossRef] [PubMed] [Google Scholar]
- Global hypomethylation in childhood asthma identified by genome-wide DNA-methylation sequencing preferentially affects enhancer regions. Allergy. 2023;78:1489-506.
- [CrossRef] [PubMed] [Google Scholar]
- Worldwide trends in the burden of asthma symptoms in school-aged children: Global Asthma Network Phase I cross-sectional study. Lancet. 2021;398:1569-80.
- [CrossRef] [PubMed] [Google Scholar]
- Longitudinal asthma phenotypes from childhood to middle-age: A population-based cohort study. Am J Respir Crit Care Med. 2023;208:132-41.
- [CrossRef] [PubMed] [Google Scholar]
- Current and optimal practices in childhood asthma monitoring among multiple international stakeholders. JAMA Netw Open. 2023;6:e2313120.
- [Google Scholar]
- Prepregnancy body mass index and risk of childhood asthma. Allergy. 2023;78:1234-44.
- [CrossRef] [PubMed] [Google Scholar]
- Early-life exposure to ambient air pollution from multiple sources and asthma incidence in children: A nationwide birth cohort study from Denmark. Environ Health Perspect. 2023;131:57003.
- [CrossRef] [PubMed] [Google Scholar]
- Asthma management and control in children, adolescents, and adults in 25 countries: A Global Asthma Network Phase I cross-sectional study. Lancet Glob Health. 2023;11:e218-28.
- [CrossRef] [Google Scholar]
- Effects of Shiwei Longdanhua formula on LPS induced airway mucus hypersecretion, cough hypersensitivity, oxidative stress and pulmonary inflammation. Biomed Pharmacother. 2023;163:114793.
- [CrossRef] [PubMed] [Google Scholar]
- LXA4 inhibits TGF-β1-induced airway smooth muscle cells proliferation and migration by suppressing the Smad/YAP pathway. Int Immunopharmacol. 2023;118:110144.
- [CrossRef] [PubMed] [Google Scholar]
- Combined epimedii folium and ligustri lucidi fructus with dexamethasone alleviate the proliferation of airway smooth muscle cells by regulating apoptosis/autophagy. J Ethnopharmacol. 2023;314:116547.
- [CrossRef] [PubMed] [Google Scholar]
- IL-13 regulates orai1 expression in human bronchial smooth muscle cells and airway remodeling in asthma mice model via LncRNA H19. J Asthma Allergy. 2022;15:1245-61.
- [CrossRef] [PubMed] [Google Scholar]
- N(6)-methyladenosine reader YTHDF1 regulates the proliferation and migration of airway smooth muscle cells through m(6)A/cyclin D1 in asthma. PeerJ. 2023;11:e14951.
- [CrossRef] [PubMed] [Google Scholar]
- Wogonin attenuates neutrophilic inflammation and airway smooth muscle proliferation through inducing caspase-dependent apoptosis and inhibiting MAPK/Akt signaling in allergic airways. Int Immunopharmacol. 2022;113:109410.
- [CrossRef] [PubMed] [Google Scholar]
- What have mechanistic studies taught us about childhood asthma? J Allergy Clin Immunol Pract. 2023;11:684-92.
- [CrossRef] [PubMed] [Google Scholar]
- Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics. 2018;19:181.
- [CrossRef] [PubMed] [Google Scholar]
- The transcription factor RFX protects MHC class II genes against epigenetic silencing by DNA methylation. J Immunol. 2009;183:2545-53.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of therapeutic targets for childhood severe asthmatics with DNA microarray. Allergol Immunopathol (Madr). 2016;44:76-82.
- [CrossRef] [PubMed] [Google Scholar]
- Transcription factor RFX7 governs a tumor suppressor network in response to p53 and stress. Nucleic Acids Res. 2021;49:7437-56.
- [CrossRef] [PubMed] [Google Scholar]
- Multi-omics analysis identifies RFX7 targets involved in tumor suppression and neuronal processes. Cell Death Discov. 2023;9:80.
- [CrossRef] [PubMed] [Google Scholar]
- The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity. Nat Immunol. 2018;19:809-20.
- [CrossRef] [PubMed] [Google Scholar]
- The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139:1478-88.
- [CrossRef] [PubMed] [Google Scholar]
- Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24:875-90.
- [CrossRef] [PubMed] [Google Scholar]
- Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J. 2013;42:65-78.
- [CrossRef] [PubMed] [Google Scholar]
- The crosstalk between autophagy and ferroptosis: What can we learn to target drug resistance in cancer? Cancer Biol Med. 2019;16:630-46.
- [CrossRef] [PubMed] [Google Scholar]
- clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-7.
- [CrossRef] [PubMed] [Google Scholar]
- Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene. 2021;40:355-68.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-370 carried by M2 macrophage-derived exosomes alleviates asthma progression through inhibiting the FGF1/MAPK/STAT1 axis. Int J Biol Sci. 2021;17:1795-807.
- [CrossRef] [PubMed] [Google Scholar]
- TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374-8.
- [CrossRef] [PubMed] [Google Scholar]
- circ_CSNK1E modulates airway smooth muscle cells proliferation and migration via miR-34a-5p/VAMP2 axis in asthma. Cell Signal. 2022;95:110340.
- [CrossRef] [PubMed] [Google Scholar]
- CircZNF652 promotes the goblet cell metaplasia by targeting the miR-452-5p/JAK2 signaling pathway in allergic airway epithelia. J Allergy Clin Immunol. 2022;150:192-203.
- [CrossRef] [PubMed] [Google Scholar]
- Antigen-specific CD4+T cells drive airway smooth muscle remodeling in experimental asthma. J Clin Invest. 2005;115:1580-9.
- [CrossRef] [PubMed] [Google Scholar]
- TIPE2 inhibits PDGF-BB-induced phenotype switching in airway smooth muscle cells through the PI3K/Akt signaling pathway. Respir Res. 2021;22:238.
- [CrossRef] [PubMed] [Google Scholar]
- Anti-IL-5 in mild asthma alters rhinovirus-induced macrophage, B-cell, and neutrophil responses (MATERIAL). A placebo-controlled, double-blind study. Am J Respir Crit Care Med. 2019;199:508-17.
- [CrossRef] [PubMed] [Google Scholar]
- Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am J Respir Crit Care Med. 2019;199:1076-85.
- [CrossRef] [PubMed] [Google Scholar]
- Loss of control of asthma following inhaled corticosteroid withdrawal is associated with increased sputum interleukin-8 and neutrophils. Chest. 2007;132:98-105.
- [CrossRef] [PubMed] [Google Scholar]
- Silencing lncRNA CDKN2BAS1 alleviates childhood asthma progression through inhibiting ZFP36 promoter methylation and promoting NR4A1 expression. Inflammation. 2023;46:700-17.
- [CrossRef] [PubMed] [Google Scholar]
- Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β and TNF-α: Receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2016;138:984-1010.
- [CrossRef] [PubMed] [Google Scholar]
- Hsa_circ_0030042 ameliorates oxidized low-density lipoprotein-induced endothelial cell injury via the MiR-616-3p/RFX7 axis. Int Heart J. 2022;63:763-72.
- [CrossRef] [PubMed] [Google Scholar]









