Translate this page into:
Targeting the Ajuba/Notch axis increases the sensitivity of colon cancer cells to 5-fluorouracil


*Corresponding authors: Xiancheng Zeng, Department of General Surgery, Guangdong Second Provincial General Hospital,466 Xingang Middle Road, Guangzhou, China. zxcq12333@163.com
Hong Wang, The First Clinical Medical College, Jinan University,601 Huangpu Avenue West Road,Guangzhou, China; Department of Gastroenterology, Guangzhou First People’s Hospital, 1 Panfu Road, Guangzhou, China. wong.hong@163.com
-
Received: ,
Accepted: ,
How to cite this article: Liang X, Liu X, Zhang L, Liu J, Yan R, Li H, et al. Targeting the Ajuba/Notch axis increases the sensitivity of colon cancer cells to 5-fluorouracil. CytoJournal. 2024;21:44. doi: 10.25259/Cytojournal_44_2024
Abstract
Objective:
Colorectal cancer is severely challenging because of the insufficient understanding of the mechanism underlying its resistance to clinical chemotherapy. The purpose of our study is to investigate the role of the LIM protein Ajuba (JUB) in the chemoresistance of colon cancer and its potential effect on clinical treatment.
Material and Methods:
The protein levels of JUB in colon cancer tissues were evaluated using Western blot analysis and immunohistochemistry assays. The correlation between JUB and the prognosis of patients with colorectal cancer was determined using Kaplan–Meier plot analysis. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were employed to determine the 50% inhibitory concentration of 5-fluorouracil (5-FU) and thus assess the effect of JUB on the effectiveness of 5-FU. In addition, the rate of cellular apoptosis was measured using fluorescence-activated cell sorting assays. Side population and sphere formation analyses were conducted to determine the role of JUB in promoting the stem cell-like traits of colon cancer cells. In vivo assays were performed and detect whether the downregulation of JUB induces 5-FU sensitivity. Moreover, luciferase and Western blot assays were employed to uncover the mechanism through which JUB promotes chemoresistance in colon cancer.
Results:
JUB expression was upregulated in chemoresistant colon cancer (P < 0.001) and correlated with relapse-free survival (P = 0.000002). Functionally, the upregulation of JUB conferred 5-FU resistance to colon cancer cells in vitro, whereas the downregulation of JUB induced 5-FU sensitivity in colon cancer cells in vivo. The high expression of JUB promoted the tumorigenic capability of colon cancer cells. Furthermore, the increased expression of JUB activated multiple downstream genes of the Notch signaling pathway with increased expression in JUB-overexpressing cells but reduced expression in JUB-silenced cells. Importantly, the inhibition of Notch signaling using a small-molecule inhibitor significantly suppressed JUB-induced chemoresistance.
Conclusion:
Results suggest that JUB plays an important role and may serve as a biomarker for the clinical treatment of patients with 5-FU-resistant colon cancer.
Keywords
Ajuba
Chemoresistance
5-fluorouracil
Notch signaling
Colon cancer
INTRODUCTION
Colon cancer is a common malignant tumor causing numerous cancer-related deaths worldwide.[1,2] The main treatment methods for colon cancer include complete mesocolon excision, systemic adjuvant chemotherapy combined with targeted drugs, surgery, and chemotherapy.[3] Chemotherapy can also be delivered as a neoadjuvant to reduce tumor size before resection.[4,5] The practicality of various chemotherapy regimens means that the overall quality of life of patients with advanced cancer has improved in recent decades. However, although the effective rate of chemotherapy can reach 50%, almost all patients with colon cancer develop drug resistance–associated chemotherapy failure.[6] Therefore, clarifying the molecular mechanism of chemoresistance and identifying molecular markers targeting chemoresistance in colon cancer are of great clinical importance for improving the prognosis of patients.
5-Fluorouracil (5-FU) is a derivative of uracil wherein the 5'-hydrogen is substituted by fluorine and is transformed into fluorouracil deoxynucleotide in cells.[7,8] With reducing folic acid as a coenzyme, fluorouracil deoxynucleotide combines with thymidine synthase to form a stable complex wherein the catalytic activity of thymidine synthase is inhibited, blocking the transformation of uracil into thymine, thereby affecting the synthesis and repair function of DNA and exerting a cytotoxic effect.[9] In addition, 5-FU can be transformed into the fluorouracil nucleoside under the action of orotate phosphoribosyltransferase. This transformation is followed by the production of fluorouracil diphosphate uridine, which is incorporated into RNA in the form of pseudometabolites, resulting in the blockade of messenger RNA (mRNA) translation and protein synthesis.[10] 5-FU chemoresistance includes primary and secondary resistance.[11,12] Primary resistance to 5-FU is caused by an increase in the mRNA and protein levels of thymidylate synthase. The causes of secondary resistance to 5-FU include the loss of enzymes that catalyze the metabolism of 5-FU to produce active products, an increase in the activity of the enzyme that catalyzes the catabolism of 5-FU, a lack of the reducing folate substrate, and the overexpression and mutation of certain genes.
The Notch signaling pathway is activated by the interaction between Notch receptors bound to the cell membrane and their ligands (such as Jagged 1–2) on parallel cells. The release of Notch intracellular domains from the plasma membrane into the nucleus, which forms complexes with CSL: CBF1/RBP-Jκ/suppressor of hairless/LAG-1) transcription factors and coactivators, can promote the transcription of Notch target genes.[13] Zhou et al.[14] found that in subcutaneous xenograft models, inhibiting the Notch1/Hes family BHLH transcription factor 1 signaling pathway can enhance the chemical sensitivity of human gastric cancer cells to 5-FU. Li et al.[15] showed that the transfection of a NOTCH1 small interfering RNA can downregulate the expression levels of multidrug resistance-associated protein 1 (MRP1). Another study demonstrated that knocking down NOTCH1 can inhibit the proliferation and invasiveness of intrahepatic cholangiocarcinoma cells and that the inhibition rate of 5-FU markedly increased.[16] Moreover, 5-FU treatment downregulated the expression levels of P-glycoprotein and MRP1 in NOTCH1-silenced cells compared with the control treatment and subsequently increased sensitivity to 5-FU in vitro.[16] In summary, the activation of the Notch1 pathway is closely related to 5-FU tolerance.
Ajuba (JUB) is a member of the adapter or scaffold protein family. It possesses a nuclear output signal that facilitates its ability to shuttle between the nucleus and cytoplasm. JUB has been reported to participate in the assembly of protein complexes that determine biological functions, such as cell fate, cytoskeleton reprogramming, mitosis control, and cell differentiation.[17,18] Several studies have demonstrated that JUB functions as a transcriptional corepressor for a subset of growth factor independent 1, which plays an important role in gene transcription regulation.[19-21] Furthermore, JUB acts as a hypoxic regulator and Von Hippel–Lindau tumor suppressor, enabling the efficient degradation of hypoxiainducible factor 1A.[22] Herein, we aim to elucidate the function and detailed mechanism of JUB in colon cancer chemoresistance.
MATERIAL AND METHODS
Cell lines
SW480 (CCL-228™) and SW620 (CCL-227™) cell lines were purchased from the American Type Culture Collection (Manassas, VA, the USA), cultured in accordance with the manufacturer’s requirements, and maintained in Roswell Park Memorial Institute 1640 medium (GIBCO, C11875500BT, Carlsbad, CA, the USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, 10099133C, Carlsbad, CA, the USA) at 37°C under 5% CO2. All cell lines were subjected to mycoplasma testing and short tandem repeat fingerprinting.
Clinical samples and immunohistochemistry
The 72 clinical tissues used in this study were all treated with chemotherapy drugs and stored in the form of paraffin-embedded sections. The clinical information for the tumor tissues, such as TNM staging (tumor, node, metastasis), is summarized in Table 1. Patients who showed signs of disease relapse or progression within 6 months of completing chemotherapy were considered chemoresistant, whereas those experiencing relapse or progression after 6 months were labeled as chemosensitive. The clinical materials used for this research were obtained with the patients’ prior consent. The research on the cancer tissue was approved by the Institutional Ethics Committee of Guangdong Second People’s Hospital (2023-KY-KZ-037-02). All the participants provided informed consent and the study design adhered to the declaration of Helsinki.
| Characteristics | No. of Cases |
|---|---|
| Age (years) | 72 |
| ≤55 | 21 |
| >55 | 51 |
| T stage | 72 |
| T1 | 5 |
| T2 | 8 |
| T3 | 53 |
| T4 | 6 |
| N stage | 72 |
| N0 | 40 |
| N1 | 23 |
| N2 | 9 |
| M stage | 72 |
| M0 | 67 |
| M1 | 5 |
| Clinical stage | 72 |
| I | 9 |
| IIa | 27 |
| IIb | 2 |
| IIc | 1 |
| IIIa | 4 |
| IIIb | 20 |
| IIIc | 4 |
| IV | 5 |
| Status (at follow-up) | 72 |
| Alive | 63 |
| Death because of cancer | 9 |
| Death because of causes other than cancer | 0 |
| New tumor | 72 |
| Yes | 15 |
| No | 57 |
| JUB expression | 72 |
| Negative | 1 |
| Positive | 71 |
| Low expression | 39 |
| High expression | 33 |
T (Tumor) represents the size and extent of the primary tumor. N (Lymph Node) represents the involvement of regional lymph nodes. M (metastasis) represents the situation of distant metastasis. JUB: Ajuba, M: Metastasis, N: Lymph node
Immunohistochemical (IHC) staining (E-IR-R217-18 mL, Elabscience, Wuhan, China) was conducted on 72 paraffin-embedded sections of colon cancer tissues using anti-JUB antibodies. The specific steps were as follows: Dewaxing: The samples on slides (5 μm) were sequentially placed in different concentrations of alcohol. Antigen repair: The slides were soaked in 3% hydrogen peroxide for 10 min then added with citric acid buffer (Macklin, C805019-100 g). Serum blocking: After the slides were cooled to room temperature, the citric acid buffer was removed. The slides were then washed twice with water and added with serum to block some non-specific sites. They were then incubated with the JUB antibody (Abcam, Cambridge, UK ab244285, 1:200) overnight in a refrigerator at 4°C and added with the secondary antibody. The slides were then washed thrice with Phosphate buffer saline (PBS) and added with 3,3’-diaminobenzidine color developer (Beyotime, P0202). The sections were rinsed with water and stained with hematoxylin. They were rinsed with water and dehydrated using a gradient alcohol series. Finally, the slides were soaked in xylene, sealed using neutral tree glue, and covered with coverslips. The slides were examined under a microscope (CK53-PT, Olympus Corporation, Japan), and the staining results were evaluated by two independent pathologists who were blinded to the clinical outcomes.
RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
By following the manufacturer’s guidelines, total RNA was isolated from cultured cells or tissue samples with Trizol reagent (15596–026, Invitrogen, Carlsbad, CA, the USA) and reverse-transcribed into complementary DNA (cDNA). Real-time qRT-PCR was then conducted on the cDNA by utilizing Synergy Brands (SYBR) Green I (FP303–01, TIANGEN, Beijing, China). Expression data were obtained on an ABI 7500 system (Applied Biosystems, Foster City, CA, the USA) and normalized to the expression levels of actin beta (ACTB) and calculated as 2−(Ct[gene]–Ct[β-actin]). The following primers were used: HES1 forward: 5'-TCAACACGACACCGGATAAAC-3' and reverse: 5'-GCCGCGAGCTATCTTTCTTCA-3'; MYC forward: 5'-GGCTCCTGGCAAAAGGTCA-3' and reverse: 5'-CTGCGTAGTTGTGCTGATGT-3'; GATA3 (GATA binding protein 3) forward: 5'-GCCCCTCATTAAGCCCAAG-3' and reverse: 5'-TTGTGGTGGTCTGACAGTTCG-3'; PTCRA (Pre T cell antigen receptor alpha) forward: 5'-AGCCCCATCTGGTTCTCAG-3' and reverse: 5'-AGGG CCATAGGTGAAGGCAT-3'; HES5 (Hes family BHLH transcription factor 5) forward: 5'-TGAAGCACAGCAAAG CCTTC-3' and reverse: 5'-TTCCCTACAGGCGAGAGGAG-3'; and β-actin forward: 5'-GCACAGAGCCTCGCCTT-3' and reverse: 5'-CCTTGCACATGCCGGAG-3'.
Western blot analysis
Proteins were extracted from the entire sample using a previously documented method.[23] First, the total protein of cells was collected using Radio Immunoprecipitation Assay lysis buffer (Thermo Fisher Scientific, 89900), and Pierce™ BCA Protein Assay (23227, ThermoFisher, the USA) was used to determine protein content. After protein quantification, sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel electrophoresis was conducted at a 10 ug concentration of whole-cell lysate. After electrophoresis, the protein was transferred to a polyvinylidene fluoride (PVDF) membrane (1620177, Bio-Rad Laboratories, Hercules, CA, the USA) in a transfer tank, and the PVDF membrane was placed in block solution (Tris-Buffered Saline Tween containing 5% skimmed milk) (P0216-1500 g, Beyotime, China) and incubated at 4°C overnight with the primary antibody. Subsequently, the PVDF membrane was incubated with the Horseradish Peroxidase-labeled secondary antibody at room temperature for 1 h. Finally, Enhanced Chemiluminescence luminescent solution (P0018FS, BeyoECL Moon, Beyotime, China) was used to detect the results using an imaging analyzer (LAS-4000 mini luminescent). Primary antibodies JUB (Abcam, ab244285, 1:500, Cambridge, England), cleaved caspase 3 (Proteintech, Wuhan, China, 25128-1-A P, 1:1000), glyceraldehyde-3-phosphate dehydrogenase (Proteintech, 10494-1-AP, 1:5000, Wuhan, China,), poly (ADP-ribose) polymerase (PARP) (Proteintech, 13371-1-AP, 1:1000 Wuhan, China,), and Notch1 (CST,#4147; 1:1000; Danvers, MA,USA). Subsequently, the membranes were reacted with labeled secondary antibodies.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
The indicated cells (SW480, SW620) were plated at the density of 2.5 × 103 cells per well in 96-well plates (Corning-3635, Corning, NY, the USA). The MTT assay was performed using an MTT assay solution kit (M1020-500, Solarbio, Beijing, China). Cell survival rate (%) was calculated as ( Optical Density [OD] experimental group/OD control group) × 100% using Microplate reader (Synergy LX, BioTek Instruments, Inc.).
Colony formation assay
A total of 1000 cells were seeded into six-well plates and cultured for 10 days. Subsequently, the colonies were fixed for 5 min with 10% formaldehyde (F864792-12 × 500 mL, Macklin, Shanghai, China) and stained with 1.0% crystal violet (HY-B0324A-500 mg, MedChemExpress, New Jersey, USA) for 30 s. The number of colonies was then counted (10 random ×100 fields per well) using a microscope (CK53-PT, Olympus Corporation, Japan). Cell counts were expressed as the mean number of cells per field of view.
Sphere formation analysis
SW480-Vector, SW480-JUB, and SW480-JUB+DAPT cells (n = 200) were seeded into 48-well ultralow attachment plates (Merck Millipore, MC96ULA20, Billerica, MA, the USA) and cultured in dulbecco’s modified eagle medium (DMEM)-F12 medium (Gibco, Grand Island, NY, the USA) supplemented with B27 (BD Biosciences, San Jose, CA, the USA), epidermal growth factor (BD Biosciences, San Jose, CA, the USA), bovine serum albumin (Sigma, Saint Louis, MO, the USA), and insulin (Sigma, Saint Louis, MO, the USA). The culture medium was refreshed every 2 days. Mammospheres were observed, photographed, and counted under a microscope (CK53-PT, Olympus Corporation, Japan).
Side population analysis
Cells were detached by utilizing trypsin and subsequently suspended at a concentration of 1 × 106 cells per mL in DMEM supplemented with 2% FBS. Thereafter, the cells were preincubated for 30 min at 37°C with or without 100 μM verapamil (Aladdin, V303890-1g, Shanghai, China). Subsequently, the samples were exposed to 5 μg/mL Hoechst 33342 dye (Hoechst 33342, S0485, Selleck, Shanghai, China) and incubated for 90 min at 37°C. The cells were then subjected to flow cytometry. Data were analyzed using Summit 5.2 software (Beckman-Coulter, Indianapolis, IN, the USA).
Annexin V assay
Annexin V assay was conducted with an apoptosis detection kit (CA1020-20T, Solarbio, Beijing, China) by following the manufacturer’s instructions. Cells were induced to undergo apoptosis in accordance with the experimental protocol and collected using centrifugation at 300 × g for 5 min. The cells were washed with PBS, gently resuspended, and counted. Subsequently, 5 × 105 cells were centrifuged and finally resuspended in 500 μL of ×1 Annexin V Binding Buffer working solution, which included Annexin V-APC and 5 propidium iodide staining solution. The cells were incubated at room temperature in the dark for 15–20 min then subjected immediately to machine testing and analysis (Flow cytometry, BD FACSCelesta). The concentration of 5-FU (F6627, Sigma, Steinheim, Germany) was 10 μM, and the treatment period was 24 h.
Luciferase assay
The promoter of the target gene was connected to the luciferase gene as the reporter gene, whose expression was assessed using a Dual-Luciferase® Reporter Assay System (E1910, Promega, Madison, WI, the USA). Changes in the fluorescence intensity of luciferase reflected the transcriptional regulatory effects of the promoter region.
Xenografted tumor model
Whether the downregulation of JUB expression contributes to 5-FU sensitivity was determined using a subcutaneously implanted tumor model. Nude mice were subcutaneously inoculated with SW620/Scramble and SW620/JUB-RNAi colon cancer cells and treated with 5-FU or the vehicle until tumors became detectable by touch. CAnN.Cg-Foxn1nu/Crl/c-nude mice (6 weeks) were purchased from the Guangdong Provincial Animal Experiment Center and randomly allocated into four groups (n = 5 per group). A total of 1 × 106 cells were injected subcutaneously into the inguinal folds of the nude mice, which were then treated with 5-FU (25 mg/kg) (HY-90006-200 mg, MedChemExpress, Monmouth Junction, NJ, the USA) every 4 days for 30 days and anesthetized using sodium pentobarbital (P3761; Sigma-Aldrich; Merck KGaA) through intraperitoneal injection (30 mg/kg). Tumors were examined once a week, and their length and width were obtained. Tumor volume was calculated as volume (mm3) = (length × width2)/2. All experimental procedures were approved by the Institutional Animal Care and Use Committee of Guangzhou Medical University (G2023-162). All animal experiments were conducted according to the animal experiment rules and ethics of the Animal Experiment Center of Guangzhou Medical University, Guangzhou, China.
Vectors and short hairpin (shRNA)
A JUB expression construct was generated using the pCDHCMV-MCS-EF1-puro retrovirus plasmid, and the human JUB shRNA construct was generated by applying the PLKO.1-U6-puro plasmid. The polymerase chain reaction-amplified primers were forward: ATGGAGCGGTTAGGAGAGAAAGC and reverse: TCACTTGTCATCGTCATCCTTG (synthesized by Invitrogen). The RNAi sequences were RNAi#1: GCAATC GCACCAGCGGCATCA and RNAi#2: GCACCTGTATCAA GTGCAACA (synthesized by Invitrogen).
Statistical analysis
The statistical analysis methods used in this study included one way anova and Student’s two-tailed t test and were performed using the Statistical Package for the Social Sciences 21.0 statistical software package (IBM Corp., Armonk, NY, the USA). A statistical significance threshold of P < 0.05 was employed in the analysis.
RESULTS
JUB is overexpressed in chemoresistant colon cancer tissues and contributes to poor prognosis
We first preformed Western blot analyses and found that compared with those in chemosensitive colon cancer tissues, JUB levels markedly increased in the five chemoresistant colon cancer tissues [Figure 1a]. We further discovered that JUB expression increased in the 33 chemoresistant colon cancer tissues that received standardized chemotherapy [Figure 1b]. Importantly, survival analysis revealed that patients with colon cancer exhibiting high JUB expression showed markedly poorer relapse-free survival than those with low JUB expression [Figure 1c]. Taken together, these findings suggest that JUB expression is upregulated in chemoresistant colon cancer and contributes to poor patient prognosis.
![Ajuba (JUB) is overexpressed in chemoresistant colon cancer tissues and contributes to poor prognosis. (a). Expression analysis of JUB expression in each group (n = 5). (b) Immunohistochemical staining indicating the JUB protein levels in each group (chemosensitive [n = 39], chemoresistant [n = 33]). (c) Kaplan–Meier plot analysis of JUB expression levels in each group (P < 0.001). Data are presented as mean ± standard deviation; ***P < 0.001. (GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, HR: Hazard ratio)](/content/105/2024/21/1/img/Cytojournal-21-44-g001.png)
- Ajuba (JUB) is overexpressed in chemoresistant colon cancer tissues and contributes to poor prognosis. (a). Expression analysis of JUB expression in each group (n = 5). (b) Immunohistochemical staining indicating the JUB protein levels in each group (chemosensitive [n = 39], chemoresistant [n = 33]). (c) Kaplan–Meier plot analysis of JUB expression levels in each group (P < 0.001). Data are presented as mean ± standard deviation; ***P < 0.001. (GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, HR: Hazard ratio)
Upregulation of JUB confers 5-FU resistance to colon cancer cells in vitro
Furthermore, the IC50 assay showed that JUB overexpression induced 5-FU resistance in colon cancer cells, whereas JUB repression (JUB-Ri1 and JUB-Ri2) lowered resistance to 5-FU compared with the control treatment [Figure 2a]. Moreover, we found that the increased expression of JUB enhanced the growth capacity of the cells, whereas the silencing of JUB (JUB-Ri1 and JUB-Ri2) had the opposite effect, as revealed by cell growth and apoptosis assays [Figure 2b and c]. In addition, the protein expression levels of activated caspase 3 and PARP notably reduced in JUB-overexpressing colon cancer cells but increased in JUB-Ri1 and JUB-Ri2 cancer cells under 5-FU treatment [Figure 2d and e].
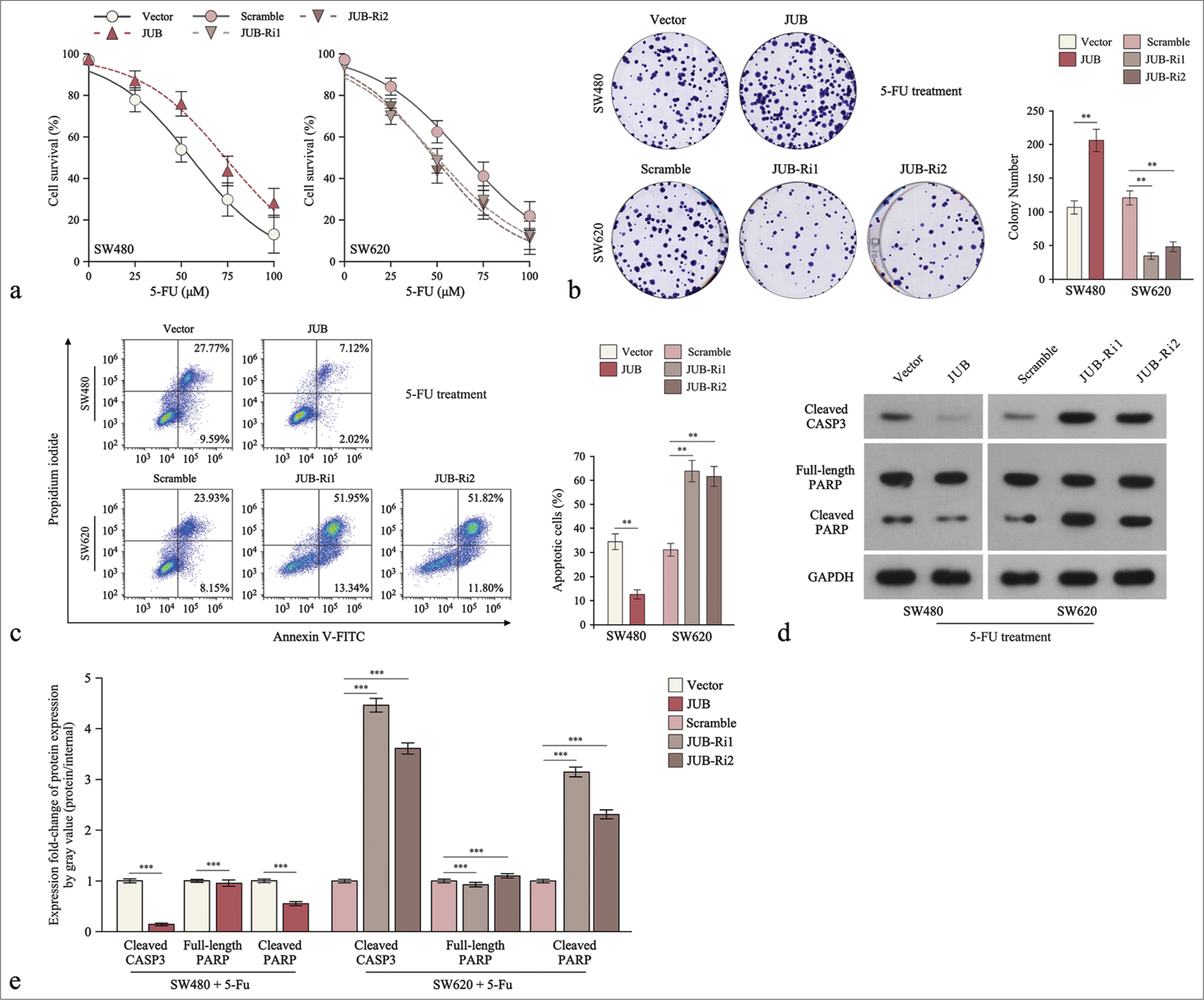
- Upregulation of Ajuba (JUB) contributes to cytotoxicity to colon cancer cells in vitro. (a) 5-fluorouracil IC50 analysis of the indicated cells. (b) Colony number in the indicated groups. (c) Analysis of the proportion of apoptotic cells in the indicated cells. (d) Levels of cleaved caspase3 and poly (ADP-ribose) polymerase (PARP) in the indicated cells. (e) Quantification of cleaved caspase3 and PARP in the indicated cells. Data are presented as the mean ± standard deviation of three independent experiments; **P < 0.01, ***P < 0.001. (5-FU: 5-Fluorouracil, GADPH: Glyceraldehyde-3-phosphate dehydrogenase, JUB-Ri1: JUB-RNAi#1, JUB-Ri2:JUB-RNAi#2, FITC: fluorescein isothiocyanate, CASP3: caspase 3)
Downregulation of JUB induces 5-FU sensitivity in colon cancer cells in vivo
Treatment with SW620/JUB-RNAi plus 5-FU induced a remarkable reduction in tumor growth [Figure 3a]. Tumor volume analysis revealed that JUB repression plus 5-FU significantly inhibited the increase in tumor weight compared with the control plus 5-FU (P < 0.05) [Figure 3b]. In tumor tissues, JUB inhibition plus 5-FU also significantly inhibited the expression of Notch1 compared with the control plus 5-FU (P < 0.001) [Figure 3c].
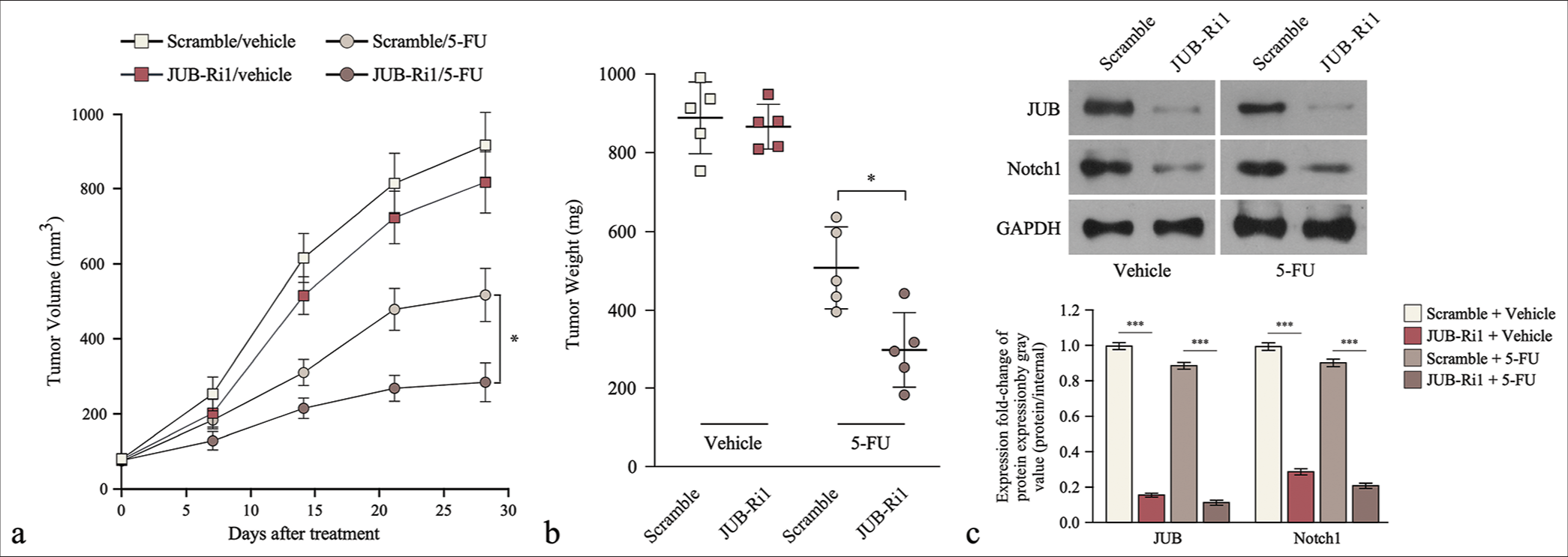
- Downregulation of Ajuba (JUB) induces 5-fluorouracil sensitivity in colon cancer cells in vivo. (a) Tumor volume (left) and (b) weight (right) at different time points in each group. (c) JUB protein levels in the indicated tumor tissues. Data are presented as mean ± standard deviation; n = 5; *P < 0.05, ***P < 0.001. (5-FU: 5-Fluorouracil, GADPH: Glyceraldehyde-3-phosphate dehydrogenase, JUB-Ri1: JUB-RNAi#1, JUB-Ri2: JUB-RNAi#2)
Upregulation of JUB promotes the stem cell-like traits of colon cancer cells
Cancer stem cells (CSCs) are the main contributors to chemoresistance. Gene Set Enrichment Analysis (GSEA) showed that JUB expression was positively correlated with stem cell-like traits [Figure 4a]. JUB-transduced cells formed more and larger tumor spheres than the control cells [Figure 4b] (P < 0.001, P < 0.001). Importantly, The Hoechst 33342 dye exclusion assay revealed that JUB significantly increased the number of side population cells among colon cancer cells (P < 0.001) [Figure 4c], suggesting that the high expression of JUB promoted the tumorigenic capability of colon cancer cells.
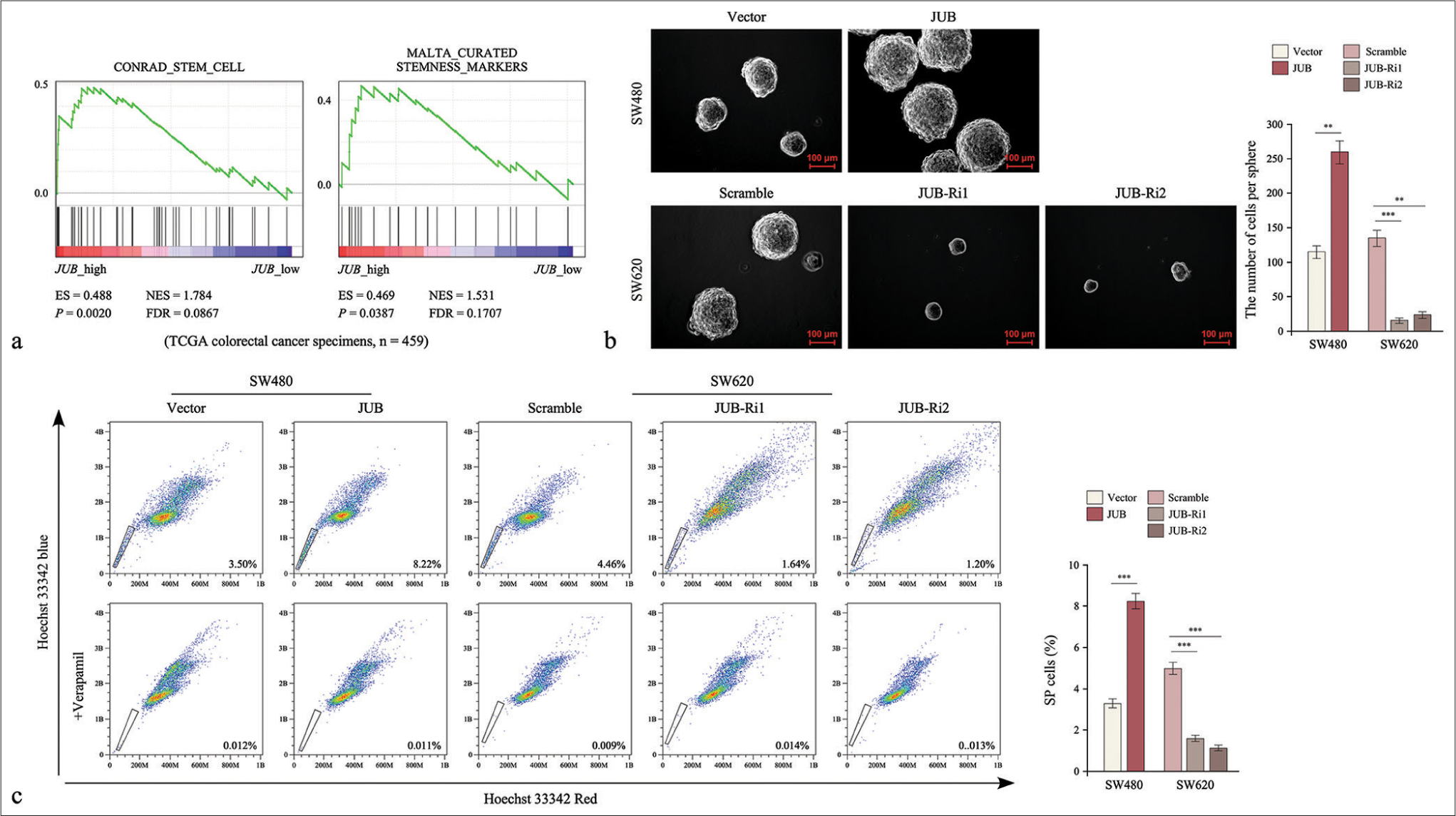
- Upregulation of Ajuba (JUB) promotes the stem cell-like traits of colon cancer cells. (a) GSEA of the correlation between JUB expression and stem cell-like trait gene expression. (b) Sphere number in the indicated groups. (c) Side population analysis on the indicated groups. Data are presented as the mean ± standard deviation of three independent experiments; n = 3, **P < 0.01, ***P < 0.001. (NES: Normalized enrichment score, FDR: False discovery rate, ES: Enrichment score, JUB: Ajuba, JUB-Ri1: JUB-RNAi#1, JUB-Ri2: JUB-RNAi#2)
Upregulation of JUB activates the notch pathway in colon cancer
We elucidated the mechanism of drug resistance induced by JUB in colon cancer. Gene Set Enrichment Analysis (GSEA)revealed that JUB expression was positively correlated with gene signatures associated with the Notch signaling pathway [Figure 5a]. Luciferase reporter activity assays showed that the activity of CSL increased in the JUB ectopic expression group but decreased in the JUB-silenced group of colon cancer cells [Figure 5b]. Moreover, the protein levels of the transmembrane fragments of NOTCH1 significantly increased in the JUB overexpression group but decreased in the JUB-silenced group of colon cancer cells (P < 0.001) [Figure 5c]. Furthermore, the qRT-PCR results demonstrated that multiple downstream genes of the Notch signaling pathway showed increased expression in JUB-overexpressing cells but reduced expression in JUB-silenced cells [Figure 5d].
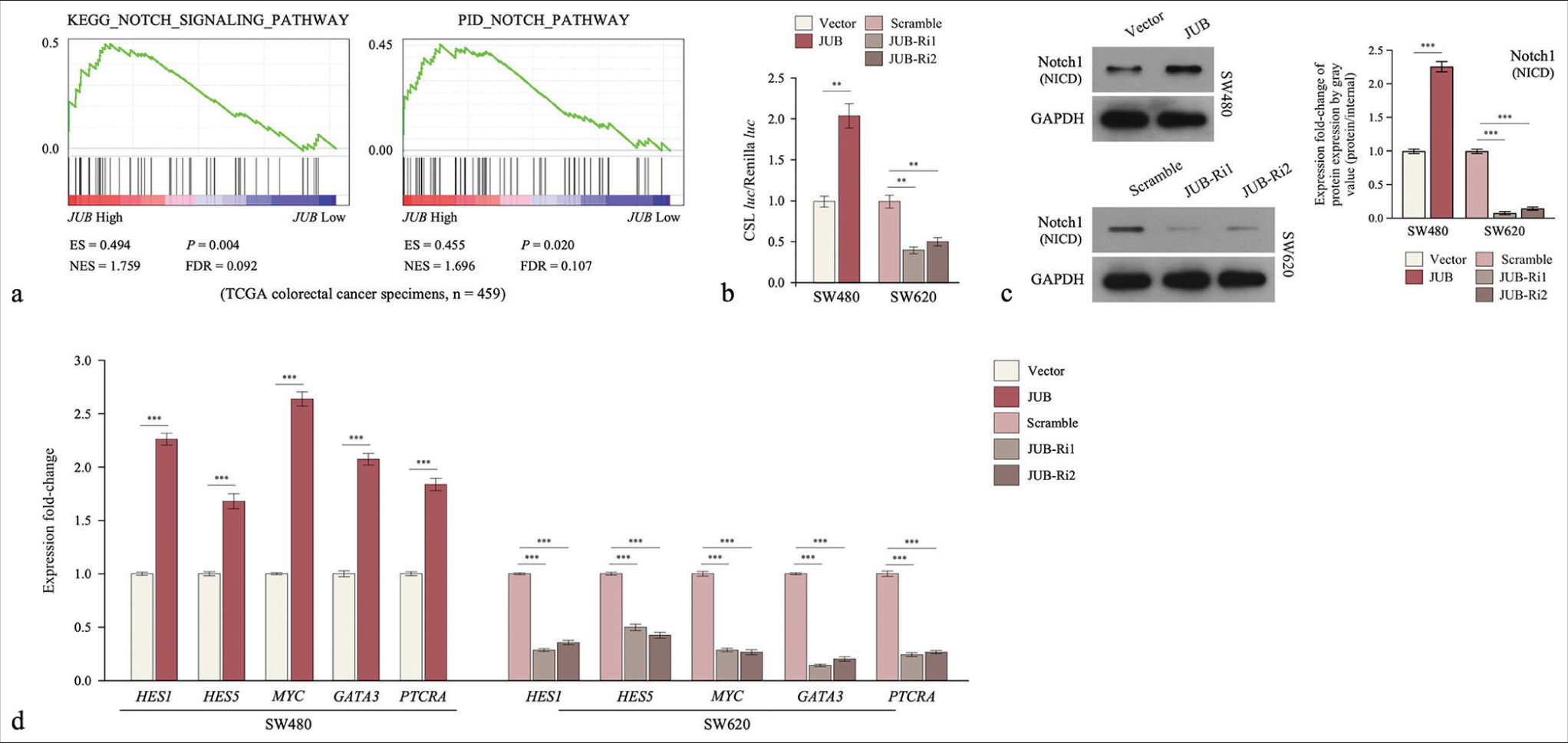
- Ajuba (JUB) upregulation activates the notch signaling pathway in colon cancer. (a) GSEA of the correlation between JUB and the notch pathway. (b) Detection of luciferase reporter activity in the indicated groups. (c) Notch 1 protein levels in the indicated cells. (d) Overlap between mRNA levels of Notch pathway gene expression and JUB-regulated gene expression. Data are presented as the mean ± standard deviation of three independent experiments; n = 3, **P < 0.01, ***P < 0.001. (ES: Enrichment score, NES: Normalized enrichment score, HES1: Hes family bHLH transcription factor 1, HES5: Hes family bHLH transcription factor 5, MYC: MYC proto-oncogene, bHLH transcription factor, GATA3: GATA binding protein 3, PTCRA: Pre T cell antigen receptor alpha, JUB: Ajuba, JUB-Ri1: JUB-RNAi#1, JUBRi2: JUB-RNAi#2, FDR: False discovery rate, GADPH: Glyceraldehyde-3-phosphate dehydrogenase, NICD: Notch1 intracellular domain)
Inhibition of the notch signaling pathway reverses JUB-induced chemoresistance
The 5-FU resistance effect of JUB on colon cancer through the Notch pathway was detected using colony formation and tumor sphere formation assays. The cells were first treated with the Notch signaling inhibitor gamma-secretase inhibitor (DAPT, 10uM, Selleck, S2215), which could remarkably reduce the expression of Notch1 but did not affect the protein level of JUB [Figure 6a]. Repressing the activity of Notch signaling significantly abrogated the drug-resistant phenotype induced by JUB in terms of the colony formation and tumor sphere formation abilities of colon cancer cells ([Figure 6b and c], P < 0.01). These results show that the Notch signaling pathway has a pivotal role in the induction of chemoresistance in colon cancer by JUB.
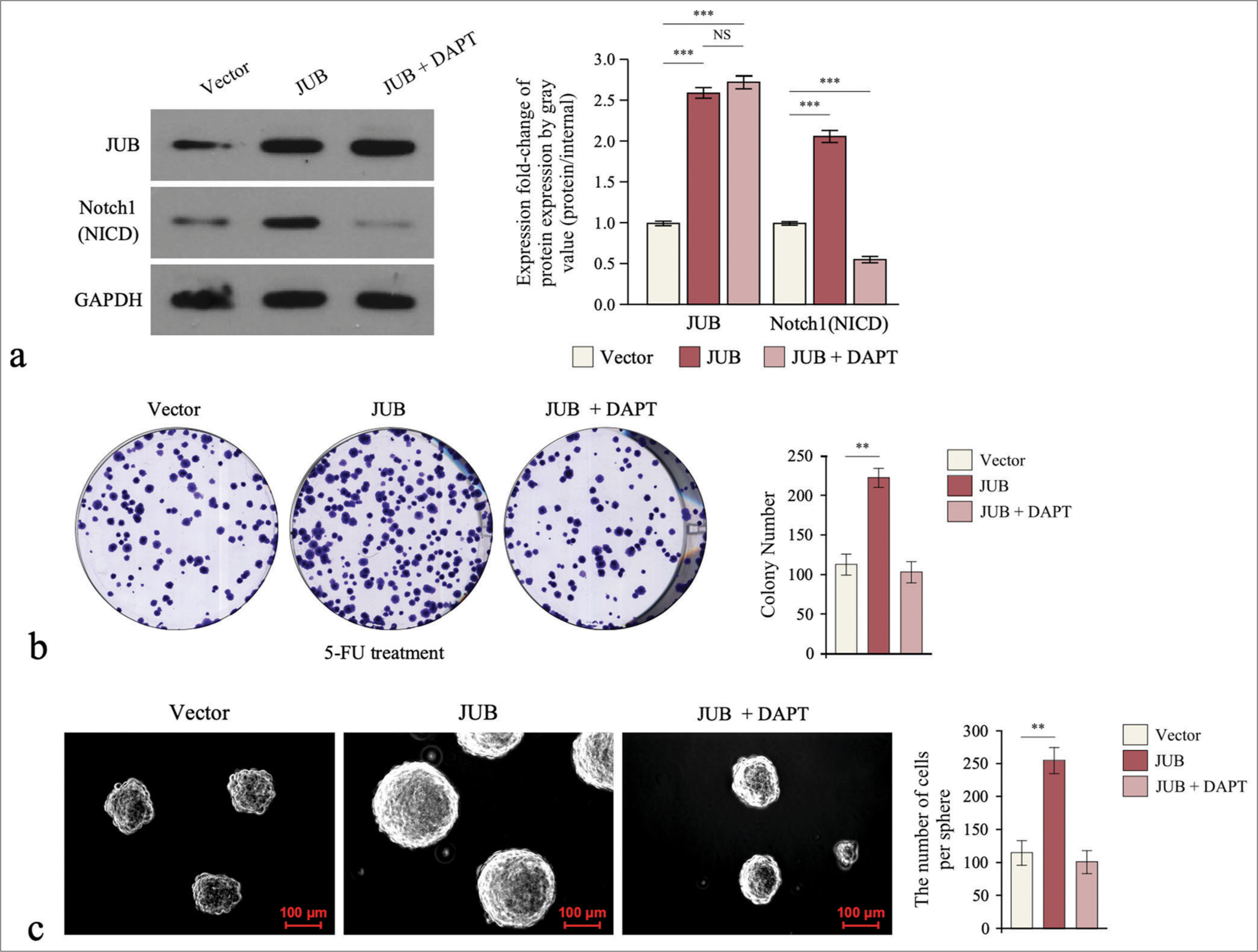
- Notch signaling pathway is required for Ajuba (JUB)-induced chemoresistance. (a) Protein levels of JUB and Notch1 (NICD) in the indicated cells. (b) Colony number in the indicated groups. (c) Sphere number in the indicated groups. Data are presented as the mean ± standard deviation SD values of three independent experiments; n = 3, **P < 0.01, ***P < 0.001. (GADPH: Glyceraldehyde-3-phosphate dehydrogenase, JUB: Ajuba, DAPT: Notch signaling inhibitor gamma-secretase inhibitor, 5-FU: 5-Fluorouracil)
DISCUSSION
Herein, we provide the first evidence for the correlation of the overexpression of JUB in chemoresistant colon cancer clinical tissues with relapse-free survival. Functionally, the overexpression of JUB conferred 5-FU resistance and promoted CSC-like phenotypes in colon cancer cells. However, the inhibition of JUB repressed this phenotype. Furthermore, the ectopic expression of JUB activated the Notch signaling pathway, and the inhibition of Notch signaling using a small-molecule inhibitor significantly suppressed JUB-induced chemoresistance.
Multiple studies have reported that JUB is overexpressed in various tumors, and the ectopic expression of JUB augments tumor metastasis, increases cisplatin resistance, and promotes cancer cell survival.[24-27] However, JUB has also been reported to be downregulated in other cancers and contributes to tumor progression. For example, Tanaka and Liu reported that JUB was downregulated in malignant mesothelioma (MM), suppressing MM cell proliferation by negatively regulating yes-associated protein activity through the large tumor suppressor kinase family; therefore, the inactivation of JUB represents a novel mechanism in MM cell proliferation.[28,29] These findings indicated that JUB acts as a pro-oncogene and tumor-suppressor gene in different types of tumor cells. Herein, we found that in colon cancer, JUB induces resistance to chemotherapy. Our results also support the view that a single gene might have different functions in different physiological and pathological states.
JUB is a member of the JUB/Zyxin subfamily of LIM proteins and functions as a transcriptional coregulator to regulate genes involved in tumor drug resistance, cell adhesion, and proliferation. It can act as a transcriptional corepressor or coactivator in transcriptional regulation in the nucleus. For example, Hou et al. reported that JUB selectively binds with 14–3–3 protein isoforms and functions as a critical cofactor for Snail-mediated transcriptional repression and epithelialmesenchymal differentiation.[30] JUB interacts with protein arginine methyltransferase 5 then binds to the N-terminal SNAG repression domain of the SNAIL transcription factor, subsequently repressing the expression of the SNAIL target gene CDH1, which encodes E-cadherin.[31] Consistently, JUB recruits p300/CBP through its LIM domain and facilitates p300/CBP binding to PPARγ to coactivate its transcriptional ability.[32] Herein, we found that JUB increased the activity of CSL and increased the expression of multiple downstream genes of the Notch signaling pathway [Figure 5b and d]. Therefore, we presume that JUB functions as a scaffold protein for the NOTCH1 intracellular domain to activate the transcription of classic Notch target genes.
FU has been an important first-line chemotherapy drug for several years.[33,34] However, the development of 5-FU resistance often hinders the good progress of chemotherapy, resulting in chemotherapy failure.[35,36] Therefore, mechanistic research and reversal of 5-FU resistance are urgently required. Drug resistance reversal can include the use of chemical drugs, gene therapy, immunomodulation, cytokine reversal, traditional Chinese medicine, and other methods.[37-39] An in-depth study of the mechanism of 5-FU resistance in gastrointestinal tumor cells can help us evaluate the efficacy of chemotherapeutic drugs in the clinic and avoid the factors causing drug resistance. This approach could actively improve the anticancer efficacy of fluorouracil drugs on the premise of reducing their toxicity and adverse reactions. Although some progress has been made, the detailed mechanisms are still unclear and require further study. In the present study, we used in vitro and in vivo assays to demonstrate that the LIM-domain protein JUB confers 5-FU resistance to colon cancer cells by activating Notch signaling. Notch signaling inhibition using a small-molecule inhibitor drastically suppressed JUB-induced 5-FU chemoresistance in colon cancer cells, suggesting that targeting the JUB/Notch axis might be a novel strategy for enhancing the response of chemoresistant colon cancer to 5-FU.
SUMMARY
We found that JUB was significantly upregulated in the clinical tissues of chemoresistant colon cancer and closely correlated with poor prognosis in patients with colon cancer. In colon cancer cells, JUB overexpression conferred 5-FU resistance and promoted CSC-like phenotypes. Furthermore, the ectopic expression of JUB activated the Notch signaling pathway and its downstream genes, thereby promoting 5-FU resistance in colon cancer. We illustrated the molecular mechanism of JUB in colon cancer chemoresistance and presented a biomarker for identifying 5-FU resistance.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
JUB: Ajuba
HRP: Horseradish Peroxidase
ECL: Enhanced Chemiluminescence
OD: Optical Density
DMEM: Dulbecco’s Modified Eagle Medium
FBS: Fetal Bovine Serum,
BALB: CAnN.Cg-Foxn1nu/Crl
GSEA: Gene Set Enrichment Analysis
5-FU:5-Fluorouracil
mRNA: messenger RNA
CSL: CBF1/RBP-Jκ/suppressor of hairless/LAG-1
PBS: Phosphate Buffer Saline
RIPA: Radio Immunoprecipitation Assay
SDS-PAGE: Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
TBST: Tris-Buffered Saline Tween
AUTHOR CONTRIBUTIONS
XHL, XCZ, and HW: Designed the study and revised and edited the manuscript; XHL, XLL, LZ, and JHL: Performed experiments, analysis, and interpretation of data; RY: Collection and assembly of data; HYL: Collected the provision of study materials or patients and and performed IHC assay; XHL, XCZ, and HW: Wrote the manuscript. All authors had approved the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The collection and use of the tissue samples were reviewed and approved by the Institutional Research Ethics Committee of Guangdong Second Provincial General Hospital (approval number: 2023-KY-KZ-037-02, Date: March 20, 2023). All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Guangzhou Medical University (approval number: G2023-162, Date: February 28, 23). The study design adhered to the declaration of Helsinki.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- NCCN guidelines insights: Colon cancer, version 2.2018. J Natl Compr Canc Netw. 2018;16:359-69.
- [CrossRef] [PubMed] [Google Scholar]
- Current standards and new trends in the primary treatment of colorectal cancer. Eur J Cancer. 2011;47(Suppl 3):S312-4.
- [CrossRef] [PubMed] [Google Scholar]
- Review article: Colorectal cancer chemotherapy. Aliment Pharmacol Ther. 2003;18:683-92.
- [CrossRef] [PubMed] [Google Scholar]
- Delaying antimalarial drug resistance with combination chemotherapy. Parassitologia. 1999;41:301-8.
- [Google Scholar]
- Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951-7.
- [CrossRef] [PubMed] [Google Scholar]
- 5-fluorouracil: Mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330-8.
- [CrossRef] [PubMed] [Google Scholar]
- 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol Ther. 2020;206:107447.
- [CrossRef] [PubMed] [Google Scholar]
- 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: Implication of DNA repair inhibition. Biomed Pharmacother. 2021;137:111285.
- [CrossRef] [PubMed] [Google Scholar]
- Computational identification of stearic acid as a potential PDK1 inhibitor and in vitro validation of stearic acid as colon cancer therapeutic in combination with 5-fluorouracil. Cancer Inform. 2021;20:11769351211065979.
- [CrossRef] [PubMed] [Google Scholar]
- Direct evidence that heterogeneity necessitates and limits the use of multidrug chemotherapy in colon cancer. Mol Med Rep. 2008;1:531-5.
- [CrossRef] [PubMed] [Google Scholar]
- Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct Target Ther. 2022;7:95.
- [CrossRef] [PubMed] [Google Scholar]
- Erteng-Sanjie capsule enhances chemosensitivity of 5-fluorouracil in tumor-bearing nude mice with gastric cancer by inhibiting Notch1/Hes1 signaling pathway. Evid Based Complement Alternat Med. 2021;2021:9980565.
- [CrossRef] [PubMed] [Google Scholar]
- Allyl isothiocyanate upregulates MRP1 expression through Notch1 signaling in human bronchial epithelial cells. Can J Physiol Pharmacol. 2020;98:324-31.
- [CrossRef] [PubMed] [Google Scholar]
- Notch1 is overexpressed in human intrahepatic cholangiocarcinoma and is associated with its proliferation, invasiveness and sensitivity to 5-fluorouracil in vitro. Oncol Rep. 2014;31:2515-24.
- [CrossRef] [PubMed] [Google Scholar]
- AJUBA: A regulator of epidermal homeostasis and cancer. Exp Dermatol. 2021;30:546-59.
- [CrossRef] [PubMed] [Google Scholar]
- Ajuba: An emerging signal transducer in oncogenesis. Pharmacol Res. 2020;151:104546.
- [CrossRef] [PubMed] [Google Scholar]
- Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585-98.
- [CrossRef] [PubMed] [Google Scholar]
- Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J Biol Chem. 2008;283:32056-65.
- [CrossRef] [PubMed] [Google Scholar]
- Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424-36.
- [CrossRef] [PubMed] [Google Scholar]
- The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat Cell Biol. 2012;14:201-8.
- [CrossRef] [PubMed] [Google Scholar]
- Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319-26.
- [CrossRef] [PubMed] [Google Scholar]
- The LIM protein AJUBA promotes colorectal cancer cell survival through suppression of JAK1/STAT1/IFIT2 network. Oncogene. 2017;36:2655-66.
- [CrossRef] [PubMed] [Google Scholar]
- The LIM protein Ajuba recruits DBC1 and CBP/p300 to acetylate ERalpha and enhances ERalpha target gene expression in breast cancer cells. Nucleic Acids Res. 2019;47:2322-35.
- [CrossRef] [PubMed] [Google Scholar]
- AJUBA increases the cisplatin resistance through hippo pathway in cervical cancer. Gene. 2018;644:148-54.
- [CrossRef] [PubMed] [Google Scholar]
- Knockout of Ajuba attenuates the growth and migration of hepatocellular carcinoma cells. Cytogenet Genome Res. 2020;160:650-8.
- [CrossRef] [PubMed] [Google Scholar]
- LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade. Oncogene. 2015;34:73-83.
- [CrossRef] [PubMed] [Google Scholar]
- Ajuba inhibits hepatocellular carcinoma cell growth via targeting of beta-catenin and YAP signaling and is regulated by E3 ligase Hakai through neddylation. J Exp Clin Cancer Res. 2018;37:165.
- [CrossRef] [PubMed] [Google Scholar]
- 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70:4385-93.
- [CrossRef] [PubMed] [Google Scholar]
- The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198-207.
- [CrossRef] [PubMed] [Google Scholar]
- The LIM protein Ajuba promotes adipogenesis by enhancing PPARγ and p300/CBP interaction. Cell Death Differ. 2016;23:158-68.
- [CrossRef] [PubMed] [Google Scholar]
- Infusional 5-FU: Historical evolution, rationale, and clinical experience. Oncology (Williston Park). 1998;12:19-22.
- [Google Scholar]
- The role of microRNAs in 5-FU resistance of colorectal cancer: Possible mechanisms. J Cell Physiol. 2019;234:2306-16.
- [CrossRef] [PubMed] [Google Scholar]
- Recent updates on mechanisms of resistance to 5-fluorouracil and reversal strategies in colon cancer treatment. Biology (Basel). 2021;10:854.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular mechanisms of drug resistance and its reversal in cancer. Crit Rev Biotechnol. 2016;36:716-26.
- [CrossRef] [PubMed] [Google Scholar]
- Resveratrol-mediated reversal of tumor multi-drug resistance. Curr Drug Metab. 2014;15:703-10.
- [CrossRef] [PubMed] [Google Scholar]
- Research progress in reversal of tumor multi-drug resistance via natural products. Anticancer Agents Med Chem. 2017;17:1466-76.
- [CrossRef] [PubMed] [Google Scholar]








