Translate this page into:
Crispr-Cas9-based long non-coding RNA interference and activation identified that the aberrant expression of Myc-regulated ST8SIA6 antisense RNA 1 promotes tumorigenesis and metastasis in hepatocellular carcinoma

*Corresponding author: Fubao Liu, Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, China. lancetlfb@126.com
-
Received: ,
Accepted: ,
How to cite this article: Liu X, Jiang D, Liu Y, Xie K, Zhao Y, Liu F. CrisprCas9-based long non-coding RNA interference and activation identified that the aberrant expression of Myc-regulated ST8SIA6 antisense RNA 1 promotes tumorigenesis and metastasis in hepatocellular carcinoma. CytoJournal. 2024;21:53. doi: 10.25259/Cytojournal_109_2024
Abstract
Objective:
Long non-coding RNAs (lncRNAs) participate in the formation, progression, and metastasis of cancer. This study aimed to explore the roles of the lncRNA ST8SIA6 antisense RNA 1 (ST8SIA6-AS1) in tumorigenesis and elucidate the underlying molecular mechanism of its upregulation in hepatocellular carcinoma (HCC).
Material and Methods:
A total of 56 in-house pairs of HCC tissues were examined, and ST8SIA6-AS1 levels were determined through real-time polymerase chain reaction (PCR). The biological behavior of ST8SIA6-AS1 by Crispr-Cas9-based gene repression and activation was determined in vitro and in vivo. The binding sites and biological behavior of Myc proto-oncogene and forkhead box A on chromatin were investigated through luciferase reporter assays, chromatin immunoprecipitation–quantitative PCR, and co-immunoprecipitation (co-IP) assays. The regulatory mechanisms of ST8SIA6-AS1 expression were analyzed with encyclopedia of DNA elements and gene expression profiling interactive analysis.
Results:
The expression of ST8SIA6-AS1 significantly increased in multiple HCC cell lines and the 56 in-house pairs of HCC tissues (P = 0.0018). Functionally, high-efficiency Crispr-Cas9-based knockdown of ST8SIA6-AS1 revealed that ST8SIA6-AS1 knockdown attenuated the proliferation, migration, and infiltration of HCC cells and considerably reduced the growth rate of subcutaneous and orthotopic HCC tumors. Conversely, ST8SIA6-AS1 overexpression considerably improved the oncogenic characteristics of the HCC cells. Furthermore, ST8SIA6-AS1 upregulation was regulated by the direct binding of transcription factor Myc to the −260 bp to+155 bp and +1003 bp to +1312 bp regions of the ST8SIA6-AS1 transcription start site, which is a segment with high level of H3K27 acetylation. Myc knockdown or treatment with the BET bromodomain inhibitor JQ-1 considerably reduced ST8SIA6-AS1 RNA expression in the HCC cells.
Conclusion:
Our study has established the oncogenic role of ST8SIA6-AS1 and elucidated the Myc-dependent upregulation mechanism of ST8SIA6-AS1 in HCC, providing a profound theoretical molecular basis for the carcinogenic function of ST8SIA6-AS1 in HCC.
Keywords
Long non-coding RNA
ST8SIA6 antisense RNA 1
Hepatocellular carcinoma
Myc proto-oncogene
Tumorigenesis
INTRODUCTION
Primary liver cancer (PLC) is a common human cancer, ranks sixth in cancer morbidity, and is the third leading cause of cancer-related deaths worldwide.[1] Hepatocellular carcinoma (HCC) is the primary type of PLC, accounting for approximately 90% of all PLC cases.[2] In the past decade, the diagnosis and treatment of HCC have remarkably improved, but due to clinically atypical symptoms, most patients are already in the middle or advanced stages of HCC when they are diagnosed.[3] In addition, the 5-year overall survival rate of patients with HCC is <30% due to the rapid progression and high incidence of tumor recurrence.[4] Therefore, constant efforts are required to elucidate the molecular mechanisms underlying the pathogenesis of HCC and identify novel diagnostic and therapeutic targets to improve its prognosis.
Non-coding RNAs (ncRNAs) comprise 98% of the transcripts of the human genome, receiving considerable interest due to their complex physiological and pathological functions.[5-7] Long ncRNAs (lncRNAs) are >200 nucleotide-long ncRNA transcripts that have no or limited protein-coding capabilities.[8] lncRNAs regulate cancer by mediating RNA–RNA, RNA–protein, or protein–protein interactions; chromatin modification; and protein modification and by sponging microRNAs (miRNAs).[5,9] To date, hundreds of lncRNAs have been found to act as tumor drivers or suppressors in various types of cancer, influencing diverse cellular malignant processes, such as cell proliferation, apoptosis, cell mitosis, migration, invasion, and drug resistance.[5,7,10-12] Hox Transcript Antisense Intergenic RNA, a well-characterized lncRNA that acts as a scaffold for histone modification complexes, is overexpressed in a broad spectrum of tumors and associated with poor prognosis and metastasis.[13] Testis Associated Oncogenic LncRNA (THOR) is a novel oncogenic lncRNA that contributes to mRNA stabilization by interacting with IGF2BP1.[14] The ectopic expression of THOR accelerates the onset of melanoma, whereas knockout of THOR in zebrafish confers resistance to melanoma onset.[15] However, current knowledge of lncRNAs involved in cancer remains limited. Here, we identified an oncogene lncRNA in HCC and named it ST8SIA6 antisense RNA 1 (ST8SIA6-AS1). Indeed, ST8SIA6-AS1 acts as a tumor driver gene that promotes the progression and metastasis of HCC.[16-21] Then, we revealed the oncogenic downstream molecular mechanism and demonstrated the crucial role of lncRNA ST8SIA6-AS1 in the occurrence and progression of HCC. Feng et al. showed that ST8SIA6-AS1 inhibits HMGA1 expression by sponging miR-142-3p, wihich accelerated LIHC cell growth while preventing cell apoptosis.[22] Another study suggested that ST8SIA6-AS1 promotes the proliferation of HCC by sponging miR-4656 and elevation of HDAC11 expression.[16] Several other studies have elaborated the other downstream regulatory and effector molecules contributing to ST8SIA6-AS1’s cancer-promoting effects, such as miR-129e5p, miR-5195-3p, and miR-338, which regulate the expression of effector molecules MAGEA3 and DCAF4L2, HOXB6, and MEPCE, respectively.[17-19] The role of ST8SIA6-AS1 in the oncological context has been extensively explored, but the precise mechanisms governing its upregulation in HCC have not been thoroughly examined. In this study, different from previous studies, we used a CRISPR-Cas9-based technology to efficiently knock down and overexpress ST8SIA6-AS1 and confirmed the promoting effect of ST8SIA6-AS1 on HCC. Importantly, we discovered that c-Myc upregulates ST8SIA6-AS1 expression by directly acting on the region near the transcription start site (TSS). This finding sheds light on the upregulation mechanism of ST8SIA6-AS1 in HCC and provides a theoretical basis for further research into its diagnostic and therapeutic potential.
MATERIAL AND METHODS
Cell lines and cell culture
HepG2 (TCHu 72), Hep3B (SCSP-5045), MHCC-97H (SCSP-5092), HCCLM3 (SCSP-5093), and HEK-293T (SCSP-502) cell lines were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). L02 and SMMC-7721 were provided by Zhongshan Hospital, China. All cell lines were verified by Short Tandem Repeats and tested negative for mycoplasma. The cell lines were cultured in dulbecco’s modified eagle medium (DMEM) (L110KJ, Yuan Pei, Shanghai China) supplemented with 10% fetal bovine serum (A2890419CP, Gibco, New York, USA) in a humidified atmosphere containing 5% CO2 at 37°C.
Clinical tissue specimens
HCC tissue specimens were obtained from patients undergoing surgical resection with a pathological diagnosis of HCC. Adjacent non-cancerous tissue specimens were collected from the same patients at least 1 cm away from the tumor margin. Patients with other malignancies or other systemic diseases were excluded from the study. The tumor tissues and their corresponding adjacent tissues were collected at the First Affiliated Hospital of Anhui Medical University. This study was approved by the Committee on Medical Ethics of the First Affiliated Hospital of Anhui Medical University and conforms to the Declaration of Helsinki.
Activation and repression of target genes using CRISPR
The genomic and promoter region sequences of human ST8SIA6-AS1 were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/gene/). Single-guide RNAs (sgRNAs) were designed in accordance with the recommendation of the Zhang Laboratory website (http://crispr.mit.edu). The sgRNAs were cloned into Lenti_gRNA-Puro (Addgene, MA, USA) in accordance with the manufacturer’s instructions.[23]
Production and infection of lentivirus
For Crispr-based ST8SIA6-AS1 overexpression, plasmids lenti dCAS9-VP64_Blast (#61425, Addgene, MA, USA), lenti MS2-P65-HSF1_Hygro (#61426, Addgene, MA, USA), and lenti sgRNA (MS2)_puro backbone (#7379, Addgene, MA, USA) with sgRNAs against the human ST8SIA6-AS1 promoter region were used. Lenti-dCas9-KRAB-blast (#89567, Addgene, MA, USA) and Lenti_gRNA-Puro (#84752, Addgene, MA, USA) with sgRNAs against the human ST8SIA6-AS1 promoter region were used to repress the expression of ST8SIA6-AS1.[24] All plasmids or plasmid backbones were obtained from Addgene.
For lentivirus production, lentiviral vectors expressing target fragments with the lentiviral packaging plasmid psPAX2 (#12260, Addgene, MA, USA) and envelope-expressing plasmid pMD2.G (#12259, Addgene, MA, USA) were simultaneously transfected into 293T cells with Lipofectamine 2000 (11668019, Thermo Fisher, MA, USA). The supernatant containing a lentivirus was harvested after 48 h of transfection. The target cell lines were infected with lentiviruses at a final polybrene concentration of 8 mg/mL for 24 h.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNA was extracted using Trizol reagent (15596018CN, Invitrogen, CA, USA), and complementary (cDNA) was synthesized using the HiScript III 1st Strand cDNA synthesis kit (R312, Vazyme, Nanjing, China). qRT-PCR was performed using ChamQ SYBR quantitative polymerase chain reaction (qPCR) Master Mix (Q331, Vazyme, Nanjing, China). The relative expression levels of lncRNAs were calculated using the comparative Ct (2−ΔΔCt), and glyceraldehyde-3-phosphate dehydrogenase was the endogenous control. All reactions were performed in accordance with the manufacturer’s instructions. All primer sequences are listed in Supplementary Table 1.
Cell cycle and apoptosis analysis
For cell cycle analysis, target cells were harvested, washed twice with phosphate buffered saline (PBS), and then treated with 70% alcohol at −20°C overnight. The cells were washed twice with PBS, and then, 200 mL of 100 mg/mL RNase I was added to the cells. The cells were incubated for 30 min at 37°C, stained with propidium iodide (PI; ST1569, Beyotime, Shanghai, China), and subjected to flow cytometry (BD LSRFortessa X-20). For apoptosis analysis, the cells were stained with annexin V and PI according to the manufacturer’s instructions (Thermo Fisher, MA, USA) and analyzed using flow cytometry for apoptosis monitoring, the analysis software is FlowJo 10 (BD, NJ, USA).
In vivo tumorigenicity assays
All animal experiments were approved by the committee on medical ethics of The First Affiliated Hospital of Anhui Medical University and conducted in accordance with the China Animal Welfare Legislation. All mice were kept in an Specific pathogen Free environment and euthanized by cervical dislocation at the end of the study. Six-week-old male athymic BALB/c nude mice were obtained from the Shanghai Slac Laboratory Animal Co. HCCLM3 cells were harvested in serum-free DMEM and subcutaneously injected into the right flank of each mouse. Then, each mouse was injected with 1 × 107 cells. The mice were euthanized at the time when tumors were 10 mm in diameter. For orthostatic liver tumor transplantation, tumors from first transplantation were dissected into approximately 2 × 2 × 2 mm in size and re-transplanted to the liver of nude mice. The tumor volumes were measured by Vernier caliper and calculated with a formula of long diameter × short diameter × short diameter × 0.5.
Statistical analysis
GraphPad Prism 8.0 software (GraphPad-Prism Software Inc., San Diego, CA) was used for conducting statistical analyses and creating graphs. Experiments were performed in triplicate and repeated at least 2 times. The data were presented as mean ± standard deviation. Analysis of variance tests, t-tests, and Tukey tests were applied to different situations. P < 0.05 indicated statistical significance (*P < 0.05, **P < 0.01, and ***P < 0.001).
RESULTS
ST8SIA6-AS1 was significantly upregulated in HCC
To explore the expression of ST8SIA6-AS1 in HCC, we first analyzed the data obtained from the public database gene expression profiling interactive analysis (GEPIA). ST8SIA6-AS1 expression was significantly upregulated in 369 liver cancer tissues and 160 paracarcinoma tissues (P < 0.05) [Figure 1a]. ST8SIA6-AS1 was limited in normal human tissues, only highly expressed in the testes, prostate, and nerves, and had low expression level in the liver (data were obtained from GEPIA) [Figures 1b, c and and Supplementary Figure 1a]. However, ST8SIA6-AS1 expression was almost specifically upregulated in HCC tissues, indicating that this lncRNA plays an important role in HCC.
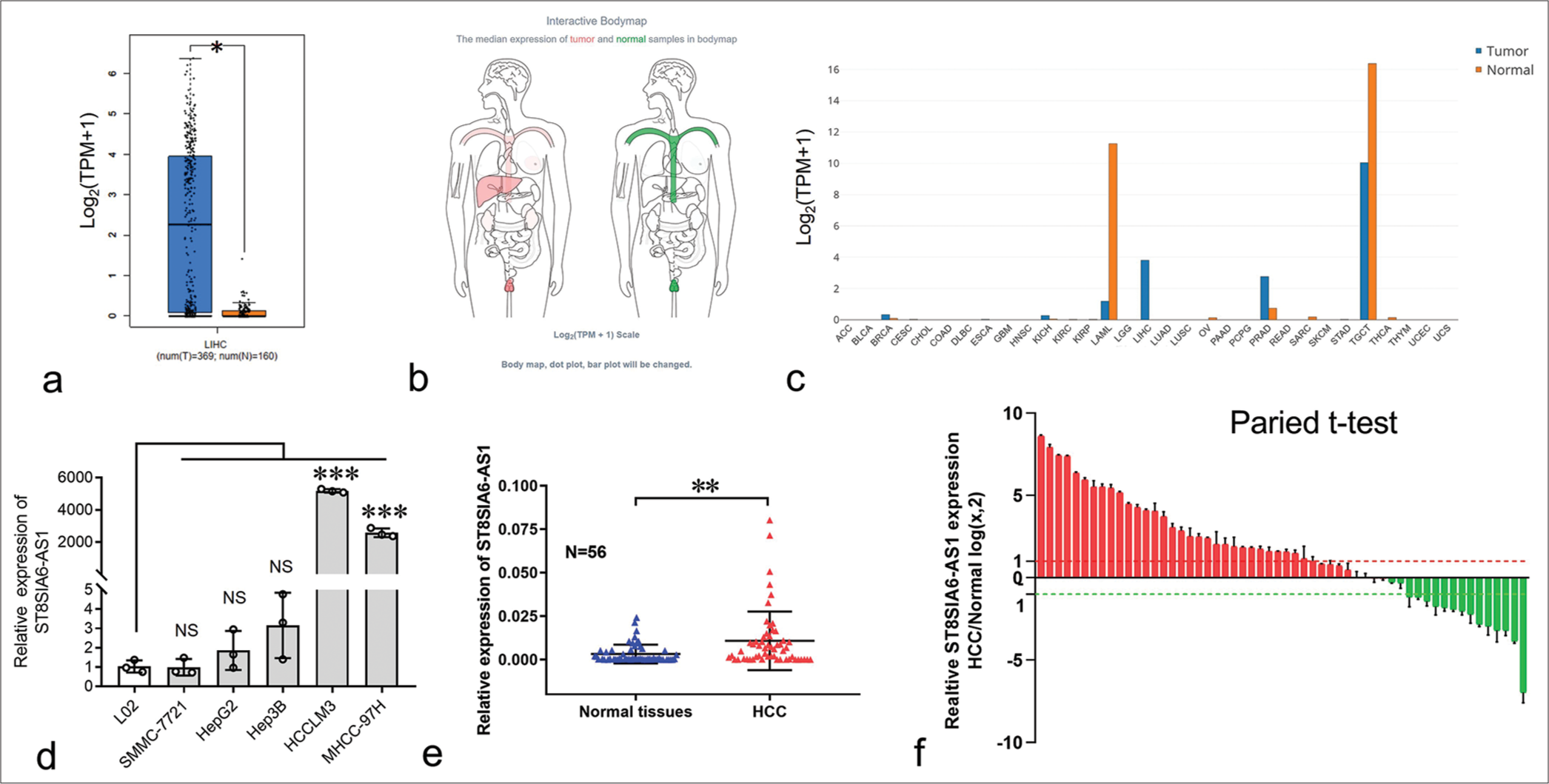
- Expression of ST8SIA6-AS1 was considerably upregulated in HCC and HCC cell lines. (a) The relative expression of ST8SIA6-AS1 in 369 HCC samples compared with 160 normal samples obtained from the GEPIA database. (b) The ST8SIA6-AS1median expression of tumor (red) and normal samples (green) in Bodymap (from the GEPIA database). (c) The ST8SIA6-AS1 expression profile across all tumor samples and corresponding normal tissues obtained from the GEPIA database. (d) The expression levels of ST8SIA6-AS1 in HCC cell lines. (e) The relative expression of ST8SIA6-AS1 in 56 in-house pairs of HCC tissues. (f) Pairwise analysis of the expression of ST8SIA6-AS1 in 56 in-house pairs of HCC tissues. Error bars are shown as mean ± SD, and ANOVA-test was used. (The red column represents the up-regulated expression of ST8SIA6-AS1 in HCC compared to the paracancer sample, while the green column represents the down-regulated expression of ST8SIA6-AS1 in HCC compared to the paracancer sample.) The asterisk indicates that the data have statistical differences, *P < 0.05, **P < 0.01, and ***P < 0.001. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, TPM: Transcripts per million, NS: no significance, LIHC: Liver hepatocellular carcinoma, HCC: Hepatocellular carcinoma, GEPIA: Gene expression profiling interactive analysis, SD: Standard deviation, ANOVA: Analysis of variance, ACC: Adrenocortical carcinoma, BLCA: Bladder Urothelial Carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and Neck squamous cell carcinoma, KICH: Kidney Chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, STES: Stomach and esophageal carcinoma, TGCT: Testicular germ cell tumors, THCA: Thyroid carcinoma, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma,UCS: Uterine carcinosarcoma).
The expression of ST8SIA6-AS1 in the normal liver cell line L02 and several HCC cell lines was measured using qPCR. L02 had the lowest ST8SIA6-AS1 RNA expression among all the analyzed cell lines [Figure 1d]. In addition, the expression of ST8SIA6-AS1 was significantly upregulated in the MHCC-97H and HCCLM3 cells (P < 0.001) [Figure 1d]. Furthermore, we used qPCR to analyze the expression of ST8SIA6-AS1 in 56 in-house pairs of liver cancer and paracarcinoma tissues. Consistent with the results from the public databases, the qPCR results showed that ST8SIA6-AS1 expression was significantly higher in the liver cancer tissues than in the paracarcinoma tissue (P = 0.0018) [Figure 1e]. Pairwise analysis results showed that ST8SIA6-AS1 expression was increased (log2FC > 1), significantly downregulated (log2FC < −1), and unchanged (−1 < log2FC < 1) in 32, 14, and 10 of the 56 pairs of tissues, respectively [Figure 1f]. This result indicated that ST8SIA6-AS1 expression was considerably upregulated in the liver tissues after carcinogenesis. Overall, ST8SIA6-AS1 expression was considerably upregulated in the HCC cells.
ST8SIA6-AS1 can promote the proliferation of HCC cells in vitro
Furthermore, we investigated the biological functions of ST8SIA6-AS1 in HCC. Small interfering RNAs (siRNAs) were used to knock down the expression of ST8SIA6-AS1 in the MHCC-97H and HCCLM3 cells. However, none of the six siRNAs used in our experiment effectively knocked down ST8SIA6-AS1 (<25% of the control group) [Supplementary Figure 1(b)]. Thus, we performed Crispr-Cas9-based gene knockdown and found that two of the three sgRNAs that bind to the ST8SIA6-AS1 promoter region efficiently knocked down the expression of ST8SIA6-AS1 in two different HCC cell lines [Figure 2a-c]. Notably, the knockdown efficiency of CrisprCas9-based gene interference (CC9i)-1 and CC9i-3 was much higher than that of siRNA (99.93% and 99.88% in MHCC-97H [P < 0.001] and 98.71% and 99.84% in HCCLM3 [P = 0.001]) and was close to the gene knockout level. Then, we generated cell proliferation curves to explore the effects of ST8SIA6-AS1 knockdown on cell proliferation. ST8SIA6-AS1 knockdown in the MHCC-97H and HCCLM3 cells effectively inhibited cell growth relative to the cell growth in the control group (cells with scrambled sgRNA) [Figure 2d and e]. To further confirm this result, we conducted a Crispr-Cas9-based cell proliferation competition assay. Briefly, cells with scrambled sgRNA or interference sgRNA were mixed in a 1:1 ratio, co-cultured, and passaged. The relative abundance of each sgRNA was detected during successive cell passages [Figure 2f]. These results show that the abundance of interference sgRNA in the MHCC-97H and HCCLM3 cells decreased with cell culture generation, which indicated that ST8SIA6-AS1 knockdown decreased the proliferation of the cells compared with the vector control [Figure 2g and h]. The result of clone formation assay performed on MHCC-97H cells showed similar proliferation inhibitory effects (P < 0.001) [Figure 2i]. In addition, we investigated the effects of ST8SIA6-AS1 knockdown on cell cycle and apoptosis. It did not significantly affect the cell cycle phase distribution and apoptosis of the cell lines [Figures 2j, Supplementary Figure 2(a) and 2(b)]. In conclusion, these results indicated that ST8SIA6-AS1 knockdown inhibited the proliferation of the HCC cells in vitro.
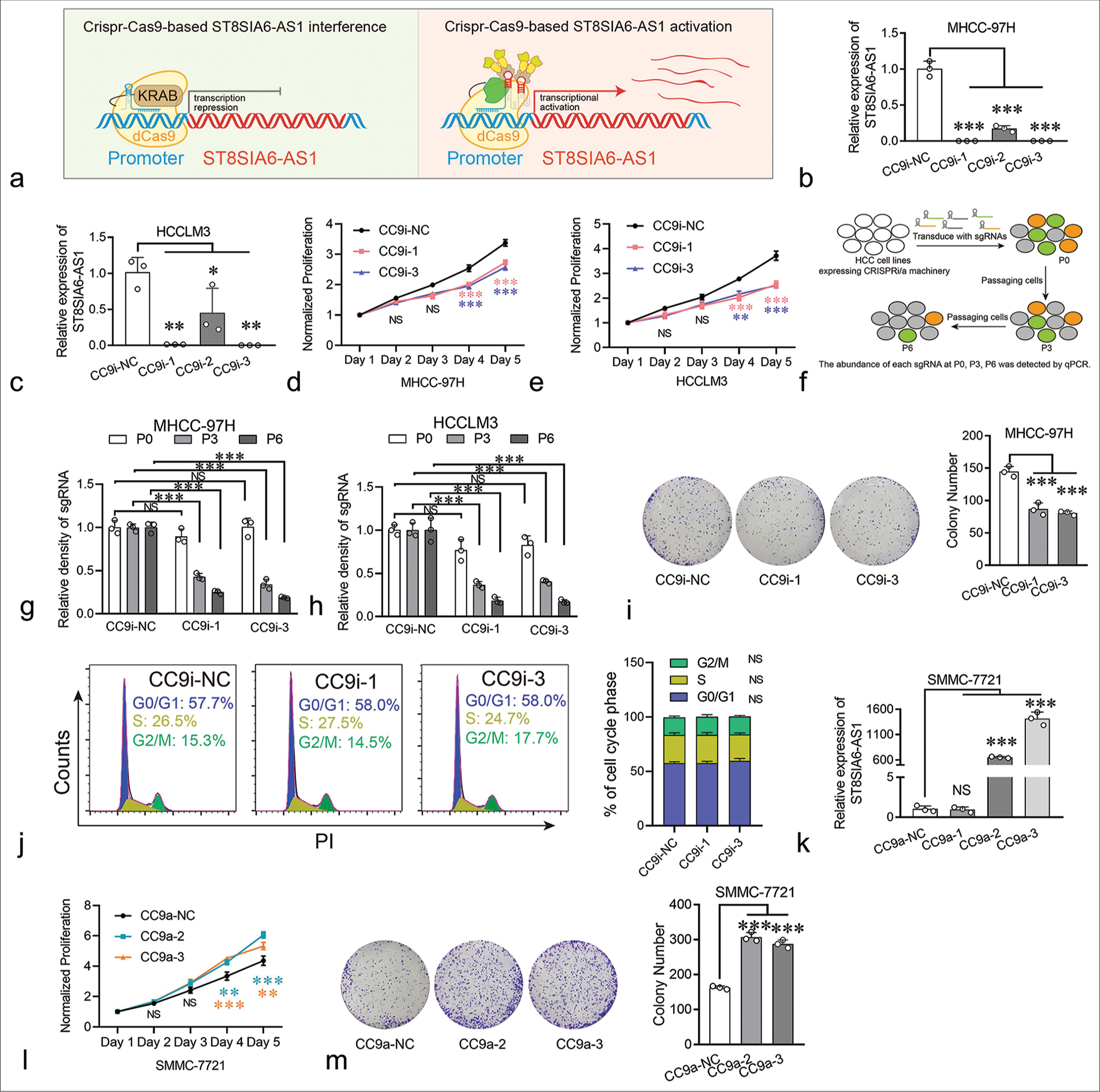
- ST8SIA6-AS1 can promote the proliferation of HCC cells in vitro. (a) Schematic illustration of Crispr-Cas9-based transcriptional silencing and activation of lncRNA ST8SIA6-AS1. This figure was independently created by the authors of this article. (b and c) CC9i efficiently downregulated the expression of ST8SIA6-AS1 in MHCC-97H and HCCLM3. (d and e) the growth curve of the HCC cell lines. (f) Experimental scheme for Crispr-Cas9-based cell proliferation competition assay (This figure was independently created by the author using Adobe Illustrator CC 2018). The abundance of sgRNAs in the HCC cell lines was detected at P0 (cell passage number), P3, and P6 through Real-time PCR. (g and h) The abundance of each sgRNA was detected through qPCR in the HCC cell lines at P0, P3, and P6. (i) Colony-forming assay was performed on MHCC-97H. The colonies were counted after 2 weeks of culture. (j) The distribution of the cell cycle phase in MHCC-97H was detected through flow cytometry. (k) CC9a efficiently upregulated the expression of ST8SIA6-AS1 in two of three sgRNAs. (l) The proliferation of SMMC-7721 was detected with a growth curve. (m) The overexpression of ST8SIA6-AS1 increased the colony-forming capacity of SMMC-7721. Error bars, mean ± S.E.M. data were analyzed with ANOVA test. The asterisk indicates that the data have statistical differences, *P < 0.05, **P < 0.01, and ***P < 0.001. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, NS: No significance, PI: Propidium iodide, CC9i: Crispr-Cas9-based gene interference, NC: Negative control, HCC: Hepatocellular carcinoma, lncRNA: Long noncoding RNAs, sgRNAs: Single-guide RNAs, PCR: polymerase chain reaction, qPCR: Quantitative polymerase chain reaction, CC9a: Crispr-Cas9-based gene activation, S.E.M.: Standard error of the mean, ANOVA: Analysis of variance.)
Furthermore, the effects of ST8SIA6-AS1 overexpression on the HCC cells were investigated. Traditional ectopic expression techniques driven by strong exogenous promoters can only express one transcript, but the number of transcripts for lncRNAs has not been fully determined. In the RefSeq database, ST8SIA6-AS1 has only one definitive transcript, whereas the LNCipedia database (https://lncipedia.org/) has annotated seven transcripts [Supplementary Table 2]. To prevent experimental bias caused by overexpression of a single transcript, we applied Crispr-Cas9-based transcriptional activation on multiple transcripts simultaneously. Results showed that two of the three sgRNAs upregulated the levels of ST8SIA6-AS1 by factors of 650 and 1415 in the SMMC-7721 cells (P < 0.001) (HCC cells with low basal expression of ST8SIA6-AS1) [Figure 2k]. The overexpression of ST8SIA6-AS1 greatly promoted the proliferation of the HCC cell lines [Figure 2l]. The result of clone formation assay for SMMC-7721 cells showed similar proliferation-promoting effects (P < 0.001) [Figure 2m]. Thus, ST8SIA6-AS1 overexpression markedly enhanced the proliferation of the HCC cell lines in vitro.
ST8SIA6-AS1 promotes the migration and infiltration of HCC cells in vitro
To evaluate the effect of ST8SIA6-AS1 on the migration and invasion of HCC cells, we performed a wound healing assay. The ST8SIA6-AS1 knockdown cell lines showed considerable reduction in migration rate, and ST8SIA6-AS1 overexpression promoted the migration of the cells [Figure 3a and b]. Transwell assay results validated these findings [Figure 3c-e]. In addition, we evaluated the effect of ST8SIA6-AS1 on the infiltration ability of the HCC cells by assessing the capacity of the cells to cross the Matrigel matrix. The results showed that ST8SIA6-AS1 knockdown considerably reduced the infiltration of the cells, whereas ST8SIA6-AS1 overexpression substantially improved the infiltration of the cells [Figure 3c-e].
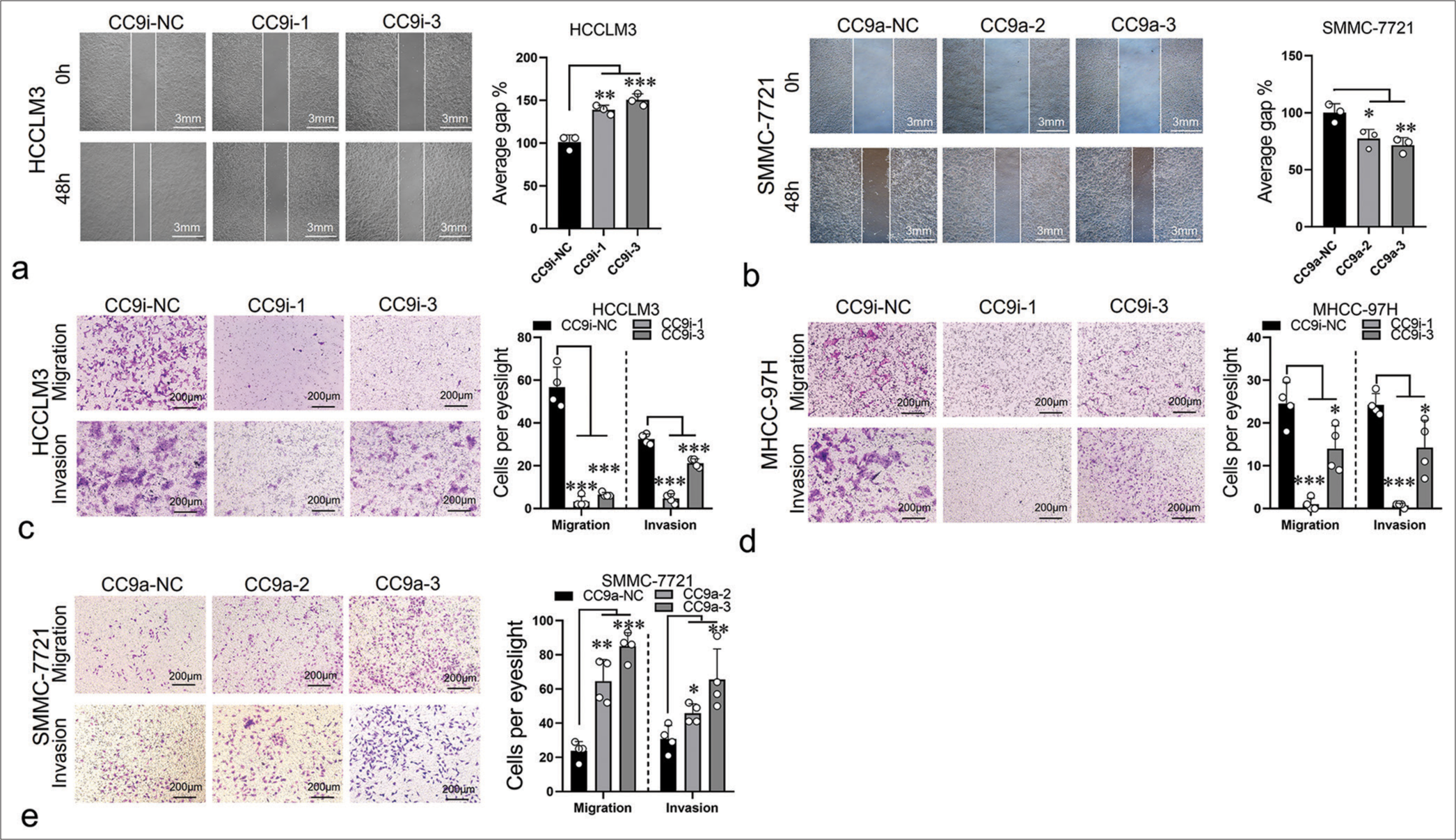
- ST8SIA6-AS1 promotes the migration and invasion of HCC cells in vitro. (a) Compared with the control group (CC9i-NC), the knockdown of ST8SIA6-AS1 (CC9i-1 and CC9i-3) decreased the migration capacity of HCCLM3. (b) Compared with the control group (CC9a-NC) the overexpression of ST8SIA6-AS1 (CC9a-2 and CC9a-3) enhanced the migration ability of SMMC-7721. (c-e) Cell migration and invasion capacity of HCCLM3, MHC-97H (c and d), and SMMC-7721 (e) were measured by Transwell migration and Matrigel invasion assays. The asterisk indicates that the data have statistical differences, *P < 0.05, **P < 0.01, and ***P < 0.001. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, HCC: Hepatocellular carcinoma, NC: Negative control, CC9a: Crispr-Cas9-based gene activation, CC9i: Crispr-Cas9-based gene interference.)
Thus, ST8SIA6-AS1 improved the oncogenic characteristics of various liver cancer cells in vitro, demonstrating its potential role in the pathogenesis of HCC.
ST8SIA6-AS1 knockdown considerably reduces the growth of liver tumor in vivo
Here, we illustrated the effects of ST8SIA6-AS1 on the proliferation, migration, and invasion of HCC cell lines in vitro and then elucidated the role of ST8SIA6-AS1 in the formation of liver tumors in vivo. We subcutaneously inoculated HCCLM3 cells with knocked down ST8SIA6-AS1 and control HCCLM3 cells into nude mice (one million cells per mouse). Tumor diameter was measured with a Vernier caliper starting from the seventh day of the experiment, and measurements were performed every 2 days. At the end of the experiment, the mice were euthanized, and tumor diameter and weight were measured [Figure 4a]. The results showed that the tumor volume of the ST8SIA6-AS1 knockdown cells was much smaller than that of the control cells (P = 0.0441) [Figure 4a and b]. We further randomly selected three tumor tissues from the experimental and control groups to detect the expression level of ST8SIA6-AS1 and determined the persistent low level of ST8SIA6-AS1 expression in the experimental groups (P < 0.001) [Figure 4c]. Immunohistochemistry revealed considerably reduced Ki-67 staining in the experimental group, indicating that ST8SIA6-AS1 knockdown reduced the proliferation of the tumor cells [Figure 4d]. Furthermore, we performed orthostatic liver tumor transplantation. These results showed that the reduction of ST8SIA6-AS1 expression markedly decelerated the growth of liver tumors in situ [Figure 4e]. In conclusion, these results demonstrated that ST8SIA6-AS1 promoted the growth of liver tumors in vivo.

- ST8SIA6-AS1 knockdown significantly reduces the growth of liver tumors in vivo. (a and b). The tumor volume of subcutaneous xenografts in nude mice injected with the HCCLM3 cells. (c) ST8SIA6-AS1 expression level was determined in the tumors of the control and ST8SIA6-AS1 knockdown groups at the end of the experiment. NC-T1, NC-T2, and NC-T3 refer to three randomly selected tumor tissues in the control group. CC9i-T1, CC9i-T2, and CC9i-T3 refer to three randomly selected tumor tissues in the ST8SIA6-AS1 knockdown group. (d) Ki67 immunohistochemical staining in xenograft tumor sections. (e) The knockdown of ST8SIA6-AS1 expression decelerated the growth of liver tumors in situ (tumor and sections of liver tissue). CC9i-NC: Control group; CC9i-3: Experimental group 3. Error bars, mean ± s.e.m. Data were analyzed with ANOVA. The asterisk indicates that the data have statistical differences, *P < 0.05, **P < 0.01, and ***P < 0.001. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, NC: Negative control, CC9i: Crispr-Cas9-based gene interference, S.E.M.: Standard error of the mean, ANOVA: Analysis of variance.)
Myc regulates the expression of ST8SIA6-AS1 by binding to its promoter region
ST8SIA6-AS1 expression was higher in liver tumors than in normal hepatocytes. We further explored the regulatory mechanism involved. ChIP-seq data from the Encylopedia of DNA Elements (ENCODE) database showed that three transcript factors, namely, Myc, Forkhead box A1 (FOXA1), and POLR2A, potentially bind to the ST8SIA6-AS1 promoter region [Figure 5a]. Myc had two binding sites (−260 bp to +155 bp and +1003 bp to +1312 bp) of the ST8SIA6-AS1 TSS [Figure 5a]. The transcription factors FOXA1 and POLR2A bound to the +1368 bp to +1489 bp and −210 bp to +193 bp restions of the TSS, respectively, indicating that these transcription factors participate in the transcriptional regulation of ST8SIA6-AS1 [Figure 5a]. In addition, overlapping MYC, FOXA1, and POLR2A binding sites, there is a significantly high H3K27 acetylation region at −960 bp to +1803 bp of the ST8SIA6-AS1 TSS, which is generally found near active regulatory elements [Figure 5a].[25] In addition, candidate cis-regulatory elements from the ENCODE mark the TSS region as a promoter-like signature.[26] This region overlaps partially with the binding sites of Myc and FOXA1 [Figure 5a]. These results indicated that the −960 bp to +1803 bp region is the transcriptional regulatory region of ST8SIA6-AS1. Myc is a critical factor in tumorigenesis and is upregulated in various tumors. FOXA1 belonging to the forkhead box family of proteins, which have conserved DNA-binding domains and play important roles in cell proliferation and HCC metastasis.[27] POLR2A RNA polymerase II subunit A is a housekeeping gene and an important component of the transcriptional complex. Therefore, in subsequent experiments, we focused on the regulatory effects of Myc and FOXA1 on ST8SIA6-AS1. We first overexpressed Myc or FOXA1 with pCDNA3.1 in SMMC-7721 cells, and the expression of ST8SIA6-AS1 was measured using qPCR. The overexpression of Myc and FOXA1 significantly increased ST8SIA6-AS1 expression compared with that in the control group (P < 0.001 and P < 0.05, respectively) [Figure 5b]. Furthermore, the knockdown of Myc or FOXA1 by siRNA in the MHCC-97H and HCCLM3 cells considerably reduced ST8SIA6-AS1 expression [Figures 5c-e and Supplementary Figure 3(a)]. Notably, Myc showed a stronger transcriptional regulation effect on ST8SIA6-AS1 than FOXA1 [Figure 5c-e]. In addition, treatment of the MHCC-97H and HCCLM3 cells with the BET bromodomain inhibitor JQ-1, which is a drug reducing Myc expression, significantly downregulated ST8SIA6-AS1 expression (P < 0.001 and P = 0.0028) [Figure 5f]. Luciferase reporter assays were performed to clarify the regulatory relationships described above. We constructed DNA fragments from the −260 bp to +1489 bp region of the ST8SIA6-AS1 TSS onto a pGL3-basic vector and transfected 293T cells simultaneously with an Myc- or FOXA1-loaded pCDNA.1 or pCDNA3.1 vector. The Myc protein significantly enhanced the luciferase signal (P < 0.001), whereas the transfection of FOXA1 did not efficiently enhance the luciferase signal. These results indicated that Myc upregulated the expression of ST8SIA6-AS1 mainly by directly binding to the ST8SIA6-AS1 region [Figure 5g]. To further determine the mode of binding between Myc and the ST8SIA6-AS1 promoter region, we performed a ChIP-qPCR assay. Compared with the control IgG, Myc showed a higher binding ability to the −260 bp to +155 bp and +1003 bp to +1312 bp regions (P = 0.0275 and P = 0.0059, respectively) [Figure 5h]. However, FOXA1 immunoprecipitation cannot effectively enrich the potential binding sites [Figure 5i]. In addition, we performed a co-IP assay to explore whether Myc and FOXA1 physically interact and found that the two proteins bind each other in an overexpression system [Figure 5j and k]. In summary, the transcription factor Myc directly bound to the ST8SIA6-AS1 promoter region at −260 bp to +155 bp and +1003 bp to +1312 bp of the TSS to upregulate the expression of ST8SIA6-AS1.
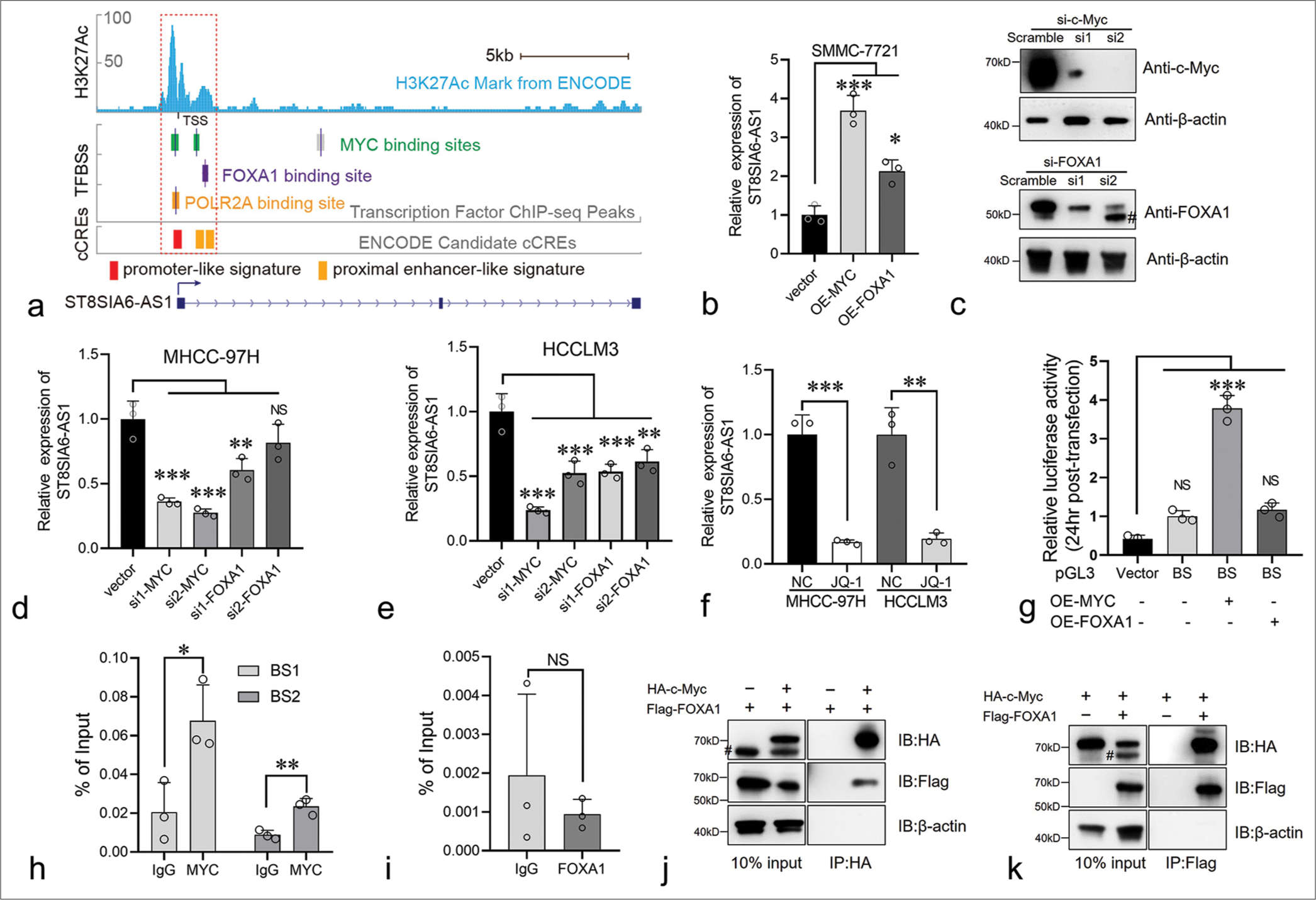
- Myc regulates the expression of ST8SIA6-AS1 by binding to its promoter region. (a) Genome browser tracks showing the Myc, FOXA1, and POLR2A binding sites in the ST8SIA6-AS1 gene locus. Data from the ENCODE database. (b) Overexpression of Myc and FOXA1 in SMMC-7721 significantly increased the RNA level of ST8SIA6-AS1. (c) MHCC-97H cells were transfected with negative control Small interference RNA (Scramble) or siRNA targeting c-Myc and FOXA1, immunoblotting analysis was performed. Asterisk, non-specific band. (d and e) SiRNA were used to knock down Myc and FOXA1 in the MHCC-97H and HCCLM3 cells, and the expression level of ST8SIA6-AS1 was detected through qPCR. (f) MHCC-97H and HCCLM3 cells with or without JQ-1 treatment for 24 h and ST8SIA6-AS1 were detected through qPCR. (g) The luciferase reporter plasmid of the binding site was co-transfected with the Myc or FOXA1-expressing plasmid in 293T. Luciferase activity was detected 24 h after transfection. Vector: Luciferase reporter vector control, BS: Plasmid with binding site region (−260 bp to +1489 bp region of the ST8SIA6-AS1 TSS). (h and i). ChIP-qPCR were performed to detect the binding of Myc or FOXA1 on the ST8SIA6-AS1 promoter region. (j and k) co-IP experiments were performed in 293T cells, and antibodies against HA and Flag tags were used. Protein levels were detected through Western blotting. Data were analyzed with Student’s t-test and ANOVA. The asterisk indicates that the data have statistical differences, *P < 0.05, **P < 0.01, and ***P < 0.001. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, TFBSs: Transcription factor binding sites, cCREs: Candidate cis-regulatory elements, OE: Overexpression, FOXA1: Forkhead box A1, qPCR: Quantitative polymerase chain reaction, BS1: Binding site 1, BS2: binding site 2, TSS: transcription start site, co-IP: Co-immunoprecipitation, ChIP-qPCR: Chromatin immunoprecipitation-quantitative polymerase chain reaction, siRNA: Small interfering RNA, HA-c-Myc: HA-tagged c-Myc, Flag-FOXA1: Flag-tagged FOXA1, #: Non-specific bands. Error bars: Mean ± Standard error of the mean, NS: No Significance, ANOVA: Analysis of variance, ENCODE: Encylopedia of DNA Elements.).
Co-expression analysis between ST8SIA6-AS1 and Myc target genes
The GEPIA database analysis showed that Myc expression was higher in the liver tumor tissues than in the paracarcinoma tissues and was consistent with the upregulated ST8SIA6-AS1 expression in the HCC tissues. However, the co-expression analysis showed no obvious co-expression relationship between Myc and ST8SIA6-AS1 [Supplementary Figure 3(b)]. Given that Myc is an upstream regulator of different oncogenes, we further analyzed the co-expression relationship between ST8SIA6-AS1 and 240 Myc target genes in liver tumors. The results showed that ST8SIA6-AS1 had a significant co-expression relationship with 208 Myc target genes (P < 0.01), of which 207 were positively related [Figure 6a]. These include several genes that have been shown to enhance the proliferation and metastasis of HCC [Figure 6b]. These results are consistent with the observed regulatory relationship between Myc and ST8SIA6-AS1 [Figure 6c].
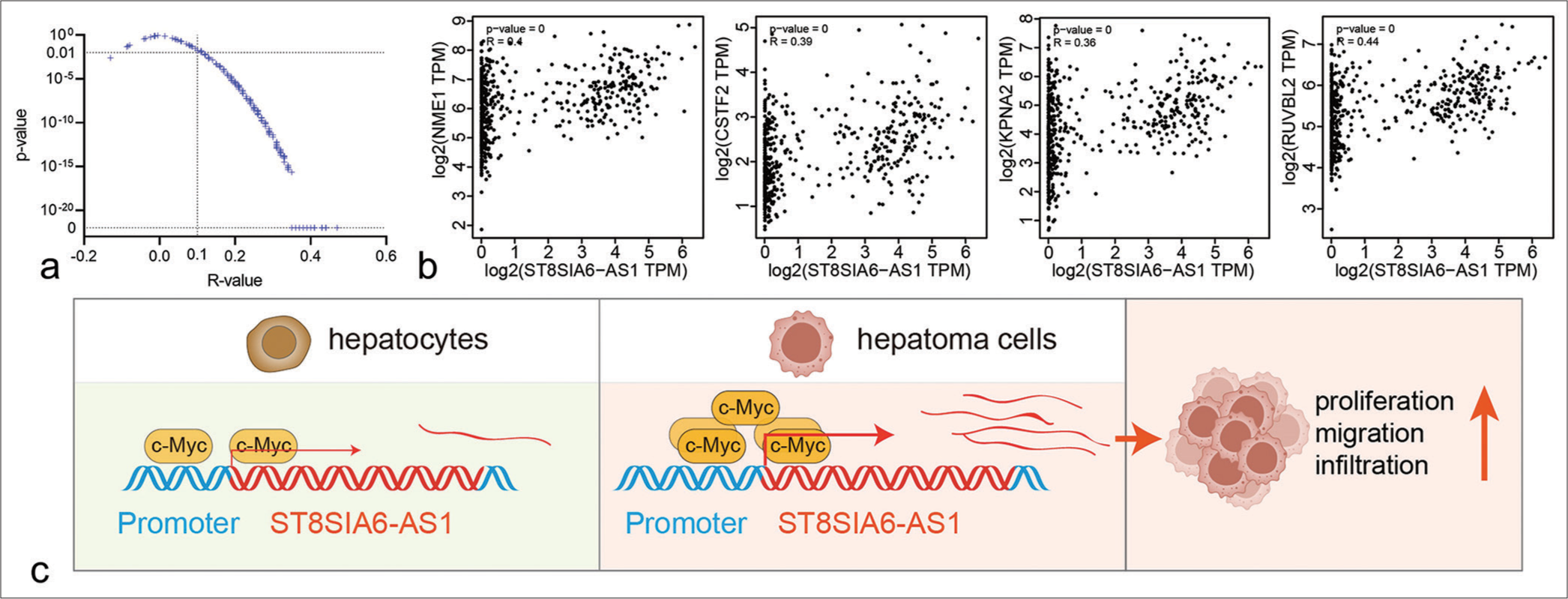
- Co-expression relationship between ST8SIA6-AS1 ang Myc target genes. (a) The scatter plot shows the correlation analysis between ST8SIA6-AS1 and 240 Myc target genes in liver tumors and GETx liver tissues (data from GEPIA). The two dotted lines refer to R = 0.1 and P = 0. (b) Representative scatter plots of co-expression between ST8SIA6-AS1 and Myc target genes. (c) Schematic of ST8SIA6-AS1 upregulation in the HCC cells. The transformation of HCC cells caused the upregulation of Myc, promoting the binding of c-Myc proteins to the promoter region, transcriptional activation of ST8SIA6-AS1, and the proliferation, migration, and invasion ability of the HCC cells. (ST8SIA6-AS1: ST8SIA6 antisense RNA 1, TPM: Transcripts per million, HCC: Hepatocellular carcinoma, GEPIA: Gene expression profiling interactive analysis.)
DISCUSSION
Many lncRNAs have been discovered through functional genomics, and the crucial roles of lncRNAs in tumorigenesis and tumor progression and metastasis have been widely studied.[28,29] Mechanistic studies have shown that lncRNAs can affect tumorigenesis, progression, and metastasis through transcriptional regulation, translation regulation, protein modification, and RNA–protein or protein–protein complex formation and participate in the regulation of important cell signaling pathways.[28] In the present study, we explored the roles of lncRNA ST8SIA6-AS1 in the initiation, progression, and transcriptional regulation mechanisms of liver tumors. ST8SIA6-AS1 is an ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 antisense RNA 1. ST8SIA6-AS1 has low expression levels in normal tissues and is mainly expressed in the testes, prostate, and nerves. Notably, it has low expression in liver cells. However, ST8SIA6-AS1 expression considerably increased after hepatocytes were transformed into hepatoma cells. This phenomenon seemed to occur only in liver and prostate cancer, demonstrating the specific role of ST8SIA6-AS1 in liver cancer. In this study, we identified the oncogenic role of ST8SIA6-AS1 and elucidated the upregulation mechanism of ST8SIA6-AS1 in HCC [Figure 6c].
We used a Crispr-Cas9-based gene knockdown and activation technology to explore the regulatory effects of lncRNA ST8SIA6-AS1 on liver tumor growth, migration, invasion, and apoptosis. Crispr-Cas9-based knockout has been widely used in mammalian coding genes, but the frameshift mutation (the most common way for CrisprCas9 to knock out coding genes) in Crispr-Cas9-based gene editing is ineffective due to the hard protein-coding capacities of lncRNAs. Therefore, one sgRNA is insufficient to cause the complete functional inactivation of lncRNAs. Fragment loss caused by dual sgRNAs is unsuitable for lncRNAs because single-exon deletion cannot effectively disrupt lncRNA function, whereas the deletion of long fragments containing multiple exons and introns greatly reduces the gene editing efficiency of Crispr-Cas9. Given the limitations of Crispr-Cas9-based gene knockout for lncRNAs, traditional siRNA-mediated gene knockdown has been preferentially used. However, the knockdown efficiency of siRNAs on lncRNAs fluctuates considerable depending on a target lncRNA. In the present study, six siRNAs were used to knock down ST8SIA6-AS1, but their efficiency was <75%. Multiple studies on ST8SIA6-AS1 have shown similar or even worse knockdown efficiency. In recent years, CrisprCas9-based gene knockdown offers obvious advantages to lncRNA research.[30,31] In the present study, the knockdown efficiency of two of the three sgRNAs in the two HCC lines was higher than 98.7%, which almost reached the level of gene knockout efficiency. The high knockdown efficiency established a premise for studying the regulatory role of ST8SIA6-AS1 in liver cancer. Crispr-Cas9-based gene activation has obvious advantages over traditional ectopic expression[32] by simultaneously activating the expression of multiple transcripts, including unannotated transcripts, and exerting minimal effects on the proportion of multiple transcripts in a physiological state. In the present study, we increased the in situ expression of ST8SIA6-AS1 650 and 1415 times.
In the present study, we determined the oncogenic role of ST8SIA6-AS1 in hepatocytes. The results showed that ST8SIA6-AS1 overexpression enhanced the proliferation, clone formation, migration, and invasion of the HCC cells. Conversely, ST8SIA6-AS1 knockdown attenuated the oncogenic characteristics of the HCC cells. In the in vivo experiments, the knockdown of ST8SIA6-AS1 greatly reduced the volume of subcutaneous tumors, reflecting a tumor-suppressive function. The knockdown of ST8SIA6-AS1 in the HCC cells in situ showed similar cancer-suppressive properties. Previous studies reported that ST8SIA6-AS1 promotes liver cancer through the adsorption of downstream miRNAs and regulation of interacting proteins. The miRNAs include miR-338-3p/NONO axis, miR-338/methylphosphate capping enzyme axis, miR-5195-3p/Homeobox B6 axis, miR-4656/The histone deacetylases 11 axis, and cancer-promoting mechanisms regulate the expression of Melanoma antigen family A 3 and DDB1-And CUL4-Associated Factor 4-Like Protein 2.[16-19,33] In the present study, we explored the main mechanism underlying ST8SIA6-AS1 upregulation in hepatocytes and found that c-Myc upregulated ST8SIA6-AS1 expression by binding to the −260 bp to +155 bp and +1003 bp to +1312 bp regions of the ST8SIA6-AS1 TSS. This conclusion was consistent with the upregulated c-Myc expression in the liver cancer cells. Moreover, the expression of ST8SIA6-AS1 in liver cancer was positively related with multiple c-Myc target genes, indirectly verifying our conclusion.
ST8SIA6-AS1 is mainly expressed in the testes and prostate under physiological conditions but not in the liver, but its expression is considerably upregulated in HCC cells. A large number of coding genes (cancer-testis antigens) are specifically expressed by the reproductive system, and their abnormal expression in non-reproductive systems can cause tumor formation.[34,35] For example, the spatiotemporal misexpression of NUT Midline Carcinoma Family Member 1 causes spindle cell sarcoma and acute lymphoblastic leukemia.[36,37] Similar studies on lncRNAs, such as ST8SIA6-AS1, are rare. Thus, some genes in lncRNAs specifically expressed by the reproductive system may be transformed into “oncogenes” when abnormally expressed in other tissues. These genes should be further explored in future studies.
Although our study has delineated the oncogenic role and upregulation mechanism of ST8SIA6-AS1 in HCC, we have not yet discerned the effects of the differential expression of ST8SIA6-AS1 on the survival of patients with HCC. The biological alterations in tumors are exceedingly complex, and ST8SIA6-AS1 may emerge as a prognostic marker in the refined subgroups of HCC. This role needs to be further explored in future studies. In addition, the upregulation of ST8SIA6-AS1 in HCC is distinct and specific, rendering it an ideal diagnostic marker for HCC. This role is particularly crucial for the early diagnosis and treatment of nascent HCC cases, potentially enhancing patient survival rates. Upregulated ST8SIA6-AS1 in patients with HCC may be released into the plasma and form free circulating RNA. PCR techniques or next-generation sequencing can be highly sensitive when detecting the elevated levels of ST8SIA6-AS1 lncRNA in the plasma and may thus considerably improve diagnostic accuracy for HCC. However, due to limitations in clinical samples, this possibility has not yet been explored. Our forthcoming research will concentrate on investigating these methods.
This article has been published as a preprint.[38]
SUMMARY
This study explored the expression and biological functions of LncRNA ST8SIA6-AS1 and the mechanisms underlying its upregulation in HCC. We demonstrated that ST8SIA6-AS1 promotes cell proliferation, migration, invasion, and enhances the tumorigenic capacity of HCC in vivo. We identified that the binding of transcription factors c-Myc and FOXA1 to the promoter region of ST8SIA6-AS1 facilitates its transcriptional activation and is a theoretical molecular mechanism for the specific transcriptional upregulation of ST8SIA6-AS1 in HCC.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
ACKNOWLEDGMENT
The authors thank all of our colleagues at Department of General Surgery of First Affiliated Hospital of Anhui Medical University for constructive discussions and technical help.
ABBREVIATIONS
lncRNAs- long noncoding RNAs
ST8SIA6- S1- ST8SIA6 antisense RNA 1
HCC- Hepatocellular carcinoma
Myc- Myc proto-oncogene
FOXA1- Forkhead box A
GEPIA-Encyclopedia of DNA Elements and Gene Expression Profiling Interactive Analysis
PLC- Primary liver cancer
ncRNAs- Noncoding RNAs
miRNAs- microRNAs
TSS- transcription start site
sgRNAs- single-guide RNAs
LIHC- Liver Hepatocellular Carcinoma
TPM-Transcripts per million
ENCODE - Encylcopedia of DNA Elements
Co-IP- Co-immunoprecipitation
ChIP-qPCR- Chromatin immunoprecipitation-quantitative polymerase chain reaction
siRNA- Small interfering RNA
NS- No Significance
ANOVA- Analysis of variance
AUTHOR CONTRIBUTIONS
XL: Conducted the experiments and wrote the manuscript; DJ: Collected the clinical samples; DJ and YL: Analyzed the data; YL, KX, and YZ: Revised the manuscript; FL: Designed the experiments and revised the manuscript. All authors reviewed, read and approved the final manuscript. All the authors contributed significantly to this study and agree with the content of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Committee on Medical Ethics of the First Affiliated Hospital of Anhui Medical University and conforms to the Declaration of Helsinki. The reference number is Quick -PJ 2023-04-29 and the date of approval is March 23, 2023. Before the patients were enrolled in this study, they were well informed about this study and were required to provide the written consents.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
Supplementary material associated with this article can be found:
FUNDING
Not applicable.
References
- Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209-49.
- [CrossRef] [PubMed] [Google Scholar]
- A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604.
- [CrossRef] [PubMed] [Google Scholar]
- Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: A systematic review and meta-analysis. Hepatology. 2018;67:422-35.
- [CrossRef] [PubMed] [Google Scholar]
- The role of non-coding RNAs in oncology. Cell. 2019;179:1033-55.
- [CrossRef] [PubMed] [Google Scholar]
- Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18.
- [CrossRef] [PubMed] [Google Scholar]
- Long non-coding RNAs in hepatocellular carcinoma: Ordering of the complicated lncRNA regulatory network and novel strategies for HCC clinical diagnosis and treatment. Pharmacol Res. 2020;158:104848.
- [CrossRef] [PubMed] [Google Scholar]
- Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96-118.
- [CrossRef] [PubMed] [Google Scholar]
- Non-coding RNAs in human cancer. Semin Cancer Biol. 2021;75:1-2.
- [CrossRef] [PubMed] [Google Scholar]
- The role of long non-coding RNA in hepatocellular carcinoma. Aging (Albany NY). 2024;16:4052-73.
- [CrossRef] [PubMed] [Google Scholar]
- Exploring non-coding RNA mechanisms in hepatocellular carcinoma: Implications for therapy and prognosis. Front Immunol. 2024;15:1400744.
- [CrossRef] [PubMed] [Google Scholar]
- HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90-7.
- [CrossRef] [PubMed] [Google Scholar]
- LncRNA THOR promotes tongue squamous cell carcinomas by stabilizing IGF2BP1 downstream targets. Biochimie. 2019;165:9-18.
- [CrossRef] [PubMed] [Google Scholar]
- Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. 2017;171:1559-72.e20.
- [CrossRef] [PubMed] [Google Scholar]
- LncRNA ST8SIA6-AS1 promotes hepatocellular carcinoma cell proliferation and resistance to apoptosis by targeting miR-4656/HDAC11 axis. Cancer Cell Int. 2020;20:232.
- [CrossRef] [PubMed] [Google Scholar]
- ST8SIA6-AS1 promotes hepatocellular carcinoma by absorbing miR-5195-3p to regulate HOXB6. Cancer Biol Ther. 2020;21:647-55.
- [CrossRef] [PubMed] [Google Scholar]
- LncRNA ST8SIA6-AS1 promotes hepatocellular carcinoma progression by regulating MAGEA3 and DCAF4L2 expression. Biochem Biophys Res Commun. 2020;533:1039-47.
- [CrossRef] [PubMed] [Google Scholar]
- Long noncoding RNA ST8SIA6AS1 promotes the migration and invasion of hypoxiatreated hepatocellular carcinoma cells through the miR338/MEPCE axis. Oncol Rep. 2021;45:73-82.
- [CrossRef] [PubMed] [Google Scholar]
- Integrated analysis of multiple transcriptomic data identifies ST8SIA6AS1 and LINC01093 as potential biomarkers in HBV associated liver cancer. Oncol Lett. 2023;25:185.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting ST8SIA6-AS1 counteracts KRAS(G12C) inhibitor resistance through abolishing the reciprocal activation of PLK1/c-Myc signaling. Exp Hematol Oncol. 2023;12:105.
- [CrossRef] [PubMed] [Google Scholar]
- ST8SIA6-AS1 contributes to hepatocellular carcinoma progression by targeting miR-142-3p/HMGA1 axis. Sci Rep. 2023;13:650.
- [CrossRef] [PubMed] [Google Scholar]
- Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281-308.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180-96.
- [CrossRef] [PubMed] [Google Scholar]
- Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931-6.
- [CrossRef] [PubMed] [Google Scholar]
- Elevated FOXA1 expression indicates poor prognosis in liver cancer due to its effects on cell proliferation and metastasis. Dis Markers. 2022;2022:3317315.
- [CrossRef] [PubMed] [Google Scholar]
- Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965-81.
- [CrossRef] [PubMed] [Google Scholar]
- The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19:77.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631-3.
- [CrossRef] [PubMed] [Google Scholar]
- CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111.
- [CrossRef] [PubMed] [Google Scholar]
- An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649-64.e20.
- [CrossRef] [PubMed] [Google Scholar]
- The HNF1a-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol Cancer. 2018;17:63.
- [CrossRef] [PubMed] [Google Scholar]
- A novel era of cancer/testis antigen in cancer immunotherapy. Int Immunopharmacol. 2021;98:107889.
- [CrossRef] [PubMed] [Google Scholar]
- Systematic identification of genes with a cancer-testis expression pattern in 19 cancer types. Nat Commun. 2016;7:10499.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging entities in NUTM1-rearranged neoplasms. Genes Chromosomes Cancer. 2020;59:375-85.
- [CrossRef] [PubMed] [Google Scholar]
- Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115:E11711-20.
- [CrossRef] [PubMed] [Google Scholar]
- Crispr-Cas9-based lncRNA interference and activation identified aberrant expression of MYC-regulated ST8SIA6-AS1 promotes tumorigenesis and metastasis in hepatocellular carcinoma. Research Square 2023
- [CrossRef] [Google Scholar]








