Translate this page into:
Detection of BRAF mutation in the cytocentrifugation supernatant fluid from fine-needle aspiration of thyroid lesions may enhance the diagnostic yield
*Corresponding author
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective:
BRAF mutations using cellular DNA from fine-needle aspiration (FNA) specimens are commonly used to support the diagnosis of papillary thyroid carcinoma (PTC). The goal of this study was to preliminarily evaluate the diagnostic utility of detecting BRAF mutations in the routinely discarded FNA specimen supernatant fluid.
Materials and Methods:
Seventy-eight FNAs of thyroid lesions were evaluated for BRAF mutations using both cellular and supernatant DNA. BRAF mutation data were correlated with cytology and surgical pathology.
Results:
Of the 78 samples evaluated, 68 (87%) had amplifiable DNA in the supernatant with 2 (3%) positive for BRAF mutations. These two samples showed no mutations in the cellular counterpart. Among the 11 samples showing morphologic findings (FNA/surgical pathology) suspicious/diagnostic of PTC, 6 (55%) samples (one supernatant and five cellulars) were positive for BRAF mutations. This suggests that testing supernatant DNA in FNA specimens may increase the diagnostic yield by 1/11 (9%) in this setting.
Conclusions:
The vast majority of routinely discarded FNA supernatants contain amplifiable DNA. In addition, profiling the mutations of BRAF and other genes using supernatant DNA may provide valuable diagnostic information to assist the diagnosis of PTC in patients with clinical/morphologic findings suspicious for malignancies and cellular DNA showing no mutations.
Keywords
BRAF
fine-needle aspirate
fine-needle aspiration supernatant
papillary thyroid carcinoma
INTRODUCTION
Worldwide, thyroid cancer is the most common endocrine malignancy making up about 1–5% of all cancers in both males and females.[12] Over the past several decades, the incidence of thyroid cancer has been reported to be increasing rapidly.[3456] Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer constituting 75–85% of all cases and therefore represents a major contributor to the worldwide increase of thyroid cancer.[5]
The BRAF mutation is the most common genetic alteration in PTC, found in 45% of cases.[7] BRAF is a cytoplasmic serine–threonine protein kinase that plays a critical role in cell signaling as an activator within the mitogen-activated protein kinase pathway. The most common BRAF mutation is a valine-to-glutamic acid change in codon 600, creating the so-called BRAF V600E mutation, which elevates the serine–threonine activity and causes a constitutive kinase signaling pathway in PTC.[8]
BRAF V600E has provided valuable prognostic information in PTC. The BRAF mutation confers a worse prognosis for PTC such as a higher risk of lymph node metastasis,[9] extrathyroidal extension,[9] advanced stage,[9] vascular invasion,[10] impaired iodine uptake,[911] recurrence,[9] and mortality.[11] In addition, PTCs that are <1 cm, but harbor BRAF mutations have been reported to be associated with adverse prognostic features such as extrathyroidal extension and lymph node metastasis, conveying a more ominous prognosis than their size would predict.[9]
BRAF V600E detection has also been a helpful diagnostic marker for PTC, and it has been shown to be a possible follow-up step to evaluate potential malignant thyroid nodules, particularly when the cytomorphological findings of the initial fine-needle aspiration (FNA) are equivocal.[1213] Studies have found that at least 20% of FNAs of thyroid nodules show indeterminate cytological findings.[7] In these cases, detection of the BRAF V600E mutation in the FNA specimen would favor a diagnosis for PTC and patients may be considered candidates for surgical intervention at higher rates than with FNA diagnosis alone.[9] Recent studies have indicated that an average of 17% of thyroid cancers, which were initially considered indeterminate on FNA biopsy, were able to be diagnosed with BRAF mutation analysis as malignant.[714151617]
To maximize the diagnostic utility of FNA specimens, the goal of the current study is to explore the diagnostic implication of detecting BRAF mutations in the supernatant fluid from cytocentrifugation of FNA specimens of thyroid lesions. The current practice for the detection of BRAF mutations is primarily done using the cellular DNA extracted from FNA specimens, while the supernatant is routinely discarded. However, at times, paucicellular or acellular FNA specimens may fail to provide adequate material for mutation testing.[18] We hypothesize that enough DNA may be present in the FNA supernatant to conduct mutation testing and that BRAF mutation analysis using supernatant may increase the diagnostic yield of the FNA.
MATERIALS AND METHODS
A total of 78 thyroid FNA samples from 72 patients (59 females, 13 males, mean: 56 years, range: 13–91 years) were used for the current study. Samples were submitted to a reference laboratory (Asuragen, Austin, TX, USA), primarily due to indeterminate cytology results, for genetic mutation profiling (including BRAF) between May 2014 and January 2015 [Table 1]. Samples were obtained, during FNA procedures performed under ultrasound guidance, from thyroid nodules. Following the FNA, some patients had thyroidectomies and corresponding surgical pathology diagnosis. The study was approved by the Institutional Review Board of Florida Hospital.
Both the cellular DNA and supernatant DNA from each individual sample were analyzed for BRAF V600E mutation. Routinely, four passes of FNA were performed during each FNA procedure. From the first three passes of the FNA, one air-dried smear and one fixed smear were made. After smear preparation, the needle was rinsed into a tube of CytoLyt solution. The needle washes in CytoLyt were centrifuged at 2000 rpm for 10 min. The supernatant was removed and a ThinPrep slide was prepared from the cells remaining in the CytoLyt tube. The supernatant was stored at 4°C until used for the current study. A fourth pass containing cellular DNA was collected directly into RNARetain media for molecular testing at the reference laboratory (Asuragen, Austin, TX, USA) using allele-specific polymerase chain reaction (PCR) with sensitivity of detecting 2% mutant BRAF alleles.[19] Of note, the Asuragen Laboratory also tested other mutations such as KRAS and NRAS in these samples. However, the results of the other mutations were not the focus of the current study and were not used for analysis.
Supernatant DNA was extracted using the Qiagen BioRobot EZ1 (Qiagen, Hilden, Germany) with 200 µL of supernatant according to the manufacturer's instruction.[20] DNA then underwent BRAF mutation testing using an in-house developed pyrosequencing-based method with a limit of 10% detection of mutant alleles. This assay has been used for more than 3 years to diagnose BRAF mutations in melanoma and colorectal cancers. PCR amplification was performed by Applied Biosystems 9700 PCR system (Applied Biosystems, Foster City, CA, USA) using primers (BRAF-F: 5’-TCT TCA TGA AGA CCT CAC AGT AAA AA-3’; BRAF-R: 5’-CCA CAA AAT GGA TCC AGA CA-3’). The amplicons were immobilized on Streptavidin Sepharose High-Performance beads (GE Healthcare Bio-Sciences, Uppsala, Sweden) as instructed by the manufacturer. Codons 594-601 of BRAF were sequenced with a sequencing primer (5’-GGA CCC ACT CCA TCG AGA-3’) in the reverse direction using PyroMark Q24 System (Qiagen, Venlo, the Netherlands).
BRAF mutation detection was then compared between samples of cellular and supernatant material. In addition, BRAF mutation data were correlated with clinical, cytology, and/or surgical pathology findings.
RESULTS
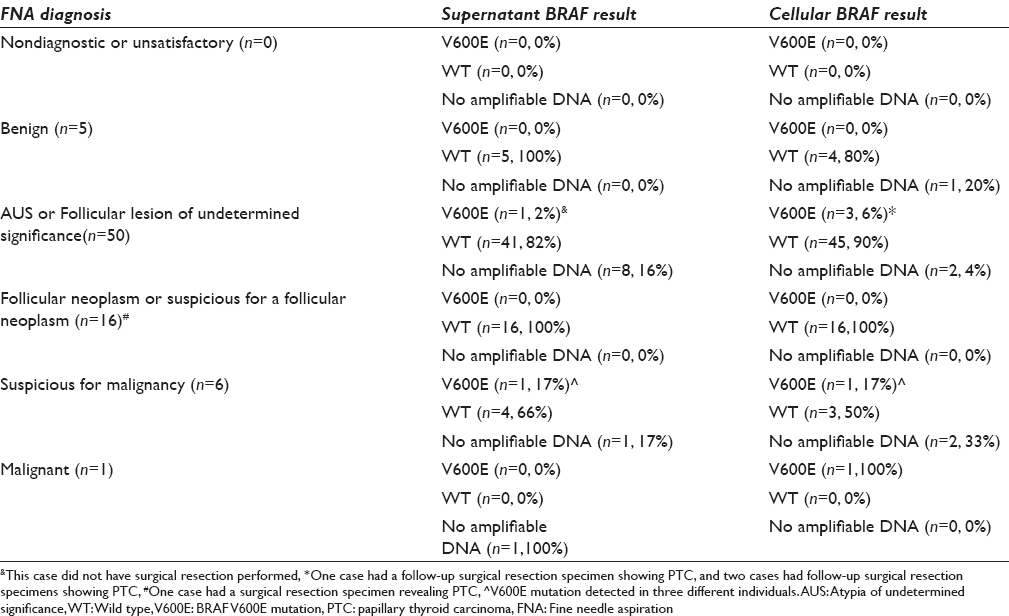
Of the 78 samples evaluated, 63 (80%) samples had amplifiable DNA from both the supernatant and cellular components for BRAF mutation analysis. About 68 (87%) samples had amplifiable DNA from the supernatant and 73 (94%) samples had amplifiable DNA from the cellular. All samples had DNA that was either amplified in the cellular or acellular supernatant [Table 1].
Cytological FNA diagnosis was rendered using the six diagnostic categories of The Bethesda System for Reporting Thyroid Cytopathology: Nondiagnostic or unsatisfactory, benign, atypia of undetermined significance (AUS) or follicular lesion of undetermined significance, follicular neoplasm or suspicious for a follicular neoplasm, suspicious for malignancy, and malignant.[21] After cytological evaluation of our samples, 0 (0%) were classified as nondiagnostic or unsatisfactory, 5 (6%) were benign, 50 (64%) were AUS or follicular lesion of undetermined significance, 16 (21%) were follicular neoplasm or suspicious for a follicular neoplasm, 6 (8%) were suspicious for malignancy, and 1 (1%) was malignant [Table 1].
Two (3%) of the 68 supernatant samples with amplifiable DNA were positive for BRAF mutations [Table 1]. Of importance, the cellular DNA counterparts of these two samples showed no BRAF V600E mutations. The first case was a 29-year-old female who had an FNA diagnosis of AUS [Figure 1a] with 27% mutant V600E allele by pyrosequencing [Figure 2b]. This case did not have follow-up surgery for further evaluation. The second case was a 52-year-old female who had an FNA showing findings suspicious for PTC [Figure 1b] which was confirmed in the resected thyroid [Figure 1c]. This case had 34% mutant V600E allele by pyrosequencing in the supernatant [Figure 2c]. These samples were checked for validity against a pyrosequenced wild type control [Figure 2a] and reagent (negative) control [Figure 2f]. In addition, the resected thyroid specimen showed the identical V600E mutation [Figure 2e] by the same method used for supernatant, confirming that the supernatant result was truly positive.

- Select fine-needle aspiration and surgical biopsies. (a) Fine-needle aspiration showing a cluster of atypical follicular cells characterized by slight nuclear enlargement and nuclear overlapping (Pap, ×400). (b) Fine-needle aspiration shows clusters of atypical follicular cells displaying slight nucleomegaly and nuclear overlapping (Diff-Quick, ×400). (c) Surgical resection reveals 1.2 cm focus of papillary thyroid carcinoma (×200)

- BRAF mutation site comparison between WT and V600E samples. (a) Wild type control; (b and c) two samples with BRAF V600E mutations in the supernatants and wild type in the cellular counterparts; (d) an example of BRAF V600E mutation in the cellular counterpart but wild-type allele in the supernatant; (e) BRAF V600E mutation as identified in the thyroidectomy specimen of the case shown in C; (f) reagent (negative) control
Five samples were positive for BRAF mutations using cellular DNA [Table 1]. Two of the samples had no amplifiable DNA in the supernatant, and the remaining three showed no mutation in the supernatant [Figure 2d]. These five samples showed AUS (n = 3), suspicious for malignancy (n = 1), and malignant PTC (n = 1) by FNA. Of note, one of the AUS cases had follow-up surgical resection specimens revealing PTC in which the V600E mutation was again confirmed by the in-house pyrosequencing assay (data not shown).
Among the eleven samples that showed FNA and/or surgical pathology findings suspicious for or diagnostic of PTC (suspicious for malignancy [n = 6], malignant [n = 1], and/or surgical pathology diagnostic of PTC [n = 4]) [Table 1], 6 (55%) samples (one supernatant and 5 cellular) were positive for the BRAF V600E mutation. This suggests that testing the supernatant DNA in FNA specimens may increase the diagnostic yield by 1/11 (9%) in this setting. However, in the setting of indeterminate cytology, the yield of testing supernatant was less and increased the BRAF mutation detection by only about 2.4% (1/42 supernatant samples with amplifiable DNA).
DISCUSSION
Our results indicate that the vast majority (87% of our samples) of routinely discarded FNA supernatants contain amplifiable DNA that could be used for molecular analysis. In addition, some samples of the supernatant contained the mutated BRAF V600E allele which was not present in the cellular counterpart of the same individual patient sample. Therefore, this routinely discarded material could be critical in some cases to diagnose and properly treat PTC patients.
In the current study, all of the BRAF V600E mutations detected in the supernatant were paired with wild-type BRAF results in the cellular component of the same FNA sample. Of interest, all patients with cellular components showing BRAF V600E mutations had the corresponding supernatants revealing either wild-type BRAF or no amplifiable DNA. The mechanisms leading to this discrepancy in results of the two components remain uncertain. The higher sensitivity (detecting 2% of mutant alleles) of the method used to detect BRAF mutations in the cellular component may have contributed to the increased detection of mutated BRAF genes in the cellular component as evaluation of the supernatant component used a method with sensitivity of detecting only 10% of alleles. Using a more sensitive method to test the supernatant, similar to the sensitivity of the one performed on the cellular component at the reference laboratory, may detect more mutations in these samples. However, this is beyond the scope of this exploratory study where the goal is to preliminarily show the diagnostic utility of the FNA supernatant.
However, the differences in sensitivity between the two different assays used cannot explain the discrepancies within the samples with mutant alleles in the supernatant and wild type alleles in the cellular component. One possibility for this discrepancy lies in the nature of the neoplastic cell population. It is conceivable that some tumors may have a higher cellular turnover (i.e., increased mitosis, apoptosis, and necrosis), resulting in the shedding of DNA (cell-free DNA)[18222324] in the interstitial fluid of the tumor, which is then caught only in the supernatant. Therefore, the lack of BRAF mutations in the cellular component of these samples could be likely due to sampling issues (i.e., cancer cells with BRAF mutations were missed by the FNA needle in the fourth pass, which is the sample submitted to the reference laboratory, resulting in a negative cellular DNA analysis while the tumor cells with the BRAF mutations were obtained from the first three FNA passes and therefore were present in the supernatant fluid leading to a positive supernatant DNA analysis). Another possibility, although less likely, is that the supernatant contained a few tumor cells that remained after centrifugation.
To the best of our knowledge, the current study represents the first attempt to use FNA thyroid specimen supernatants to test for BRAF mutations in PTC. Our findings of detecting BRAF mutations in FNA supernatants of PTC are in agreement with the results of a recent study, which demonstrated that the use of FNA supernatant specimens from patients with pancreatic cancers for molecular testing may outperform cellular DNA analysis in certain situations.[18] We attempted to quantify the DNA amounts in the supernatant; however, the majority of samples showed the amount of DNA was below the linear range of NanoDrop 1000 (Thermo Scientific, Wilmington, DE) even though 87% of our samples had enough amplifiable DNA for the detection of BRAF mutation. Of note, we only used 200 µl of about 25–50 ml supernatant for DNA extraction; therefore the yield of DNA could potentially be increased if a greater amount of supernatant was used for DNA extraction.
The percentage of BRAF mutations among AUS cases in the current study is lower than in the literature (8% or 4/50 combining both supernatant and cellular samples vs. the 15–20% found in other studies).[714151617] However, this BRAF mutation rate is compatible to that of a recent study using residual ThinPrep material from thyroid FNAs for BRAF testing.[25] The mechanisms leading to these discrepancies are unknown. Nevertheless, this does not impact the diagnostic utility of FNA supernatants.
CONCLUSIONS
Our results indicate that profiling the mutations of BRAF and other genes using DNA from routinely discarded FNA supernatants may provide valuable diagnostic information to assist with the diagnosis of PTC in a substantial percentage of patients, particularly if clinical/morphologic findings are suspicious for malignancies and no mutations are identified in cellular DNA samples. In the current study, testing supernatant DNA in the FNA specimens increases the diagnostic yield by 1/11 (9%). Based on the findings of the current study, we suggest that the FNA supernatants should be kept until the cellular molecular testing results are available. If the molecular test results using the cellular DNA are inconclusive (i.e., negative or no amplifiable DNA), the supernatants can be then submitted for molecular testing to maximize the diagnostic yield of an FNA when clinically indicated. This may prevent patients from being subjected to unnecessary further work-ups such as repeat FNA procedures.
COMPETING INTERESTS STATEMENT BY ALL AUTHORS
The authors do not have competing interests.
AUTHORSHIP STATEMENT BY ALL AUTHORS
AB and KL organized the data and wrote the draft of the paper. GC performed the mutation study. AT and MH made the diagnosis and provided the photographs. CC planed the study and wrote the final paper.
ETHICS STATEMENT BY ALL AUTHORS
The study was approved by Institutional Review Board, Florida Hospital.
LIST OF ABBREVIATIONS (In alphabetic order)
AUS: Atypia of undetermined significance
DNA: Deoxyribonucleic acid
FNA: Fine-needle aspiration
PCR: Polymerase chain reaction
PTC: Papillary thyroid carcinoma
WT: Wild type genotype.
EDITORIAL/PEER-REVIEW STATEMENT
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through automatic online system.
ACKNOWLEDGMENTS
The authors would like to thank the Molecular Diagnostic Lab staff and Cytology Lab staff at Florida Hospital Orlando, for their technology support.
REFERENCES
- Cancer Incidence in Five Continents. Vol IX. Lyon: IARC Scienfic Publications, IARC; 2007. p. :160.
- SEER Cancer Statistics Review, 1975-2012. Bethesda, MD: National Cancer Institute; 2015.
- Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA. 2006;295:2164-7.
- [Google Scholar]
- Increased incidence of thyroid carcinoma in france: A true epidemic or thyroid nodule management effects? Report from the French Thyroid Cancer Committee. Thyroid. 2004;14:1056-60.
- [Google Scholar]
- Worldwide increasing incidence of thyroid cancer: Update on epidemiology and risk factors. J Cancer Epidemiol 2013 2013 965212
- [Google Scholar]
- International patterns and trends in thyroid cancer incidence, 1973-2002. Cancer Causes Control. 2009;20:525-31.
- [Google Scholar]
- An update on molecular biology of thyroid cancers. Crit Rev Oncol Hematol. 2014;90:233-52.
- [Google Scholar]
- Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309:1493-501.
- [Google Scholar]
- Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19:1167-214.
- [Google Scholar]
- American Association of Clinical Endocrinologists, Associazione Medici Endocrinologi, and European Thyroid Association medical guidelines for clinical practice for the diagnosis and management of thyroid nodules: Executive summary of recommendations. J Endocrinol Invest. 2010;33(5 Suppl):51-6.
- [Google Scholar]
- Detection of BRAF mutation on fine needle aspiration biopsy specimens: A new diagnostic tool for papillary thyroid cancer. J Clin Endocrinol Metab. 2004;89:2867-72.
- [Google Scholar]
- Mutational analysis of BRAF in fine needle aspiration biopsies of the thyroid: A potential application for the preoperative assessment of thyroid nodules. Clin Cancer Res. 2004;10:2761-5.
- [Google Scholar]
- Analysis of BRAF point mutation and RET/PTC rearrangement refines the fine-needle aspiration diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. 2004;89:5175-80.
- [Google Scholar]
- A rapid and simple detection method for the BRAF (T1796A) mutation in fine-needle aspirated thyroid carcinoma cells. Thyroid. 2004;14:910-5.
- [Google Scholar]
- The value of mutational profiling of the cytocentrifugation supernatant fluid from fine-needle aspiration of pancreatic solid mass lesions. Mod Pathol. 2014;27:594-601.
- [Google Scholar]
- A multiplex technology platform for the rapid analysis of clinically actionable genetic alterations and validation for BRAF p.V600E detection in 1549 cytologic and histologic specimens. Arch Pathol Lab Med. 2014;138:371-8.
- [Google Scholar]
- Qiagen. EZ1 virus handbook (January). 2011.
- The Bethesda system for reporting thyroid cytopathology. Am J Clin Pathol. 2009;132:658-65.
- [Google Scholar]
- Circulating tumour-derived DNA and RNA markers in blood: A tool for early detection, diagnostics, and follow-up? Lung Cancer. 2005;49:1-12.
- [Google Scholar]
- Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J. 2010;4:11-21.
- [Google Scholar]
- Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell-free tumor DNA as indicator of breast cancer progression. Clin Cancer Res. 2012;18:5719-30.
- [Google Scholar]
- Molecular analysis of residual ThinPrep material from thyroid FNAs increases diagnostic sensitivity. Cancer Cytopathol. 2015;123:356-61.
- [Google Scholar]








