Translate this page into:
Exploration of immunocytochemical biomarkers related to central lymph node metastasis in papillary thyroid microcarcinoma

*Corresponding author: Wenhao Ren, Key laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of pathology, Peking University Cancer Hospital and Institute, Beijing, China. renwenhao@bjmu.edu.cn
-
Received: ,
Accepted: ,
How to cite this article: Rong L, Wang J, Wang Q, Zhu Y, Ren W. Exploration of immunocytochemical biomarkers related to central lymph node metastasis in papillary thyroid microcarcinoma. CytoJournal. 2025;22:18. doi: 10.25259/Cytojournal_162_2024
Abstract
Objective
The presence of central lymph node metastasis (CLNM) represents a critical determinant in ascertaining the necessity for surgical intervention in patients with papillary thyroid microcarcinoma (PTMC). However, the predominant current methodologies for confirming the central lymph node status in clinical practice are hampered by the low predictive accuracy of preoperative ultrasound examination and the high risk of preoperative fine needle aspiration (FNA). Consequently, the objective of this study is to investigate and identify specific immunocytochemical biomarkers for predicting CLNM in PTMC patients based on preoperative thyroid FNA samples.
Material and Methods
In this study, the messenger ribonucleic acid sequencing data of pathological tumor stage 1 (pT1) papillary thyroid carcinoma (PTC) accompanied by pathological node stage information were initially retrieved from The Cancer Genome Atlas database. The differential expression genes (DEGs) between the pT1N1-PTC group and the pT1N0-PTC group were ascertained through bioinformatics methodology. Subsequently, these DEGs were imported into Cytoscape software to identify hub genes. Ultimately, immunohistochemical and immunocytochemical staining were employed to validate whether the biomarkers corresponding to the main hub genes demonstrated statistical significance in predicting CLNM within propensity score-matched PTMC samples.
Results
In this study, a total of 292 DEGs and 10 hub genes were successfully identified. Subsequently, immunohistochemical and immunocytochemical staining were conducted on 208 PTMC cases selected through propensity score matching. Among these 208 cases, the biomarkers (Cytokeratin 5/6 [CK5/6], Chromogranin A [CgA], and Pair box gene 2 [Pax-2]) corresponding to the main hub genes (Cytokeratin 5 [KRT5], Cytokeratin 6 [KRT6A], Chromogranin A [CHGA], and PAX2) were subjected to immunohistochemical staining in postoperative thyroidectomy specimens, the immunohistochemical staining results revealed a statistically significant difference in CK5/6 expression between PTMCs with and without CLNM (P = 0.002). Subsequently, CK5/6 immunocytochemical staining performed on preoperative thyroid FNA liquid-based samples further corroborated that CK5/6 expression was more prone to being positive in PTMCs with CLNM (P = 0.010).
Conclusion
CK5/6 is a valuable immunocytochemical biomarker capable of predicting the occurrence of CLNM in PTMC patients prior to surgery.
Keywords
Immunocytochemistry
Keratin-5
Lymphatic metastasis
Papillary thyroid microcarcinoma
INTRODUCTION
Thyroid cancer is undergoing rapid growth as a malignant tumor on a global scale. Specifically, papillary thyroid microcarcinoma (PTMC), which is defined as papillary thyroid cancer (PTC) with a maximal tumor diameter not exceeding 1 cm, accounts for almost 50% of newly diagnosed cases of thyroid cancer.[1]
Extensive literature has indicated that certain PTMCs consistently persist in a subclinical stage, exhibit a favorable prognosis, and can even coexist with the tumor; these PTMCs with good clinical outcomes are termed low-risk PTMCs.[2] Although the criteria differ among various countries and regions, PTMCs that fulfill the criteria of no lymph node metastasis (LNM), no distant metastasis, no extrathyroidal invasion, no pathological high-risk subtype, and no tracheal or recurrent laryngeal nerve invasion are commonly defined as low-risk PTMCs.[3] Low-risk PTMCs can be managed through active surveillance, ablation therapy, or surgery. Active surveillance and ablation therapy can circumvent surgical and post-operative complications, thereby significantly enhancing the quality of life for these patients.[4] However, many countries and regions primarily choose surgical intervention when dealing with low-risk PTMCs.[5] One of the major factors is that the existing clinical examination techniques in numerous countries are incapable of accurately determining whether the cervical lymph nodes have metastasized prior to treatment. PTMC cases demonstrate a low incidence of extrathyroidal invasion, high-risk pathological subtypes, tracheal or recurrent laryngeal nerve invasion, and distant metastasis, while PTMC cases have a substantial incidence of cervical LNM. Therefore, based on the definition of low-risk PTMC, the key determinant for classifying PTMC as low-risk is whether there are cervical LNMs.[3] Nevertheless, the two main clinical assessment methods for preoperative cervical LNM have certain limitations: ultrasound examination has low accuracy in predicting LNM, especially in some institutions with poor medical standards; fine needle aspiration (FNA) examination is highly prone to accidentally injure large blood vessels and nerves when puncturing the central lymph nodes, and the puncture risk is extremely high.[6] Consequently, if doctors follow international recommendations and administer active surveillance or ablation treatment to PTMC patients who show no clinically detectable LNM (cN0), there is a risk of delaying treatment for many patients who actually have LNM.
However, the approach of relying on surgical treatment sometimes results in unnecessary surgeries for numerous PTMC patients who do not actually have LNMs. A precise diagnosis is of utmost importance for precise treatment. Hence, it is vital to emphasize the need for new techniques in the medical setting to precisely determine the preoperative condition of cervical lymph nodes. This will permit the accurate diagnosis of low-risk PTMC and subsequently facilitate the implementation of tailored and personalized treatment, thus enhancing the patient’s chances of recovery and overall quality of life.
Employing clinical variables to predict PTMC cervical LNMs is a frequently employed strategy; however, its prediction accuracy is relatively low.[7-9] The genetic information of tumor cells serves as the foundation for their biological functioning, and the advancement of high-throughput sequencing and bioinformatics technologies has made the investigation of molecular biomarkers a highly prospective approach for addressing this issue.
In our previous study,[10] we discovered that the presence of squamous cell components in PTCs was correlated with a poor prognosis. Based on this finding, we postulated that biomarkers related to squamous cell components might be linked to central LNM (CLNM) in PTMC. Based on this hypothesis, this study aims to utilize bioinformatics analysis in conjunction with immunohistochemistry (immunocytochemistry) validation to investigate and confirm biomarkers associated with squamous cell components or other biomarkers with potential predictive value for CLNM in PTMC, thereby providing a reference for the clinical management of PTMC patients.
MATERIAL AND METHODS
Screening of hub genes related to LNM
The messenger ribonucleic acid (mRNA) microarray dataset and clinical information of PTC cases were extracted from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/, National Cancer Institute, Bethesda, Maryland, USA). Subsequently, cases with a pathological tumor stage of pT1 (≤2 cm) and pathological node stage were selected and screened. The screened cases were divided into two groups based on their pathological node stage for further bioinformatics analysis: The group with LNM (pT1N1 PTC group) and the group without LNM (pT1N0 PTC group).
Differential expression genes (DEGs) in the pT1N0 PTC group and the pT1N1 PTC group were identified using the Limma package of the R software version 4.1.3 (R Foundation for Statistical Computing, Vienna, Austria). Genes from each sample that fulfilled the following criteria were preserved: (1) a |log2 (foldchange)| ≥2 and (2) a false discovery rate <0.05. The integrated dysregulated gene lists were saved for subsequent analysis. The ggplot2 and heatmap software tools were employed to generate heatmap and volcano plots of DEGs, respectively.
The DEGs were annotated with the “ClusterProfiler” Bioconductor package (version 4.4.4, Guangchuang Yu, Guangzhou, China), which combines biological data and analysis tools to offer a comprehensive set of functional annotations for genes.
To predict protein-protein interaction (PPI) networks, the Search Tool for the Retrieval of Interacting Genes (STRING)/Proteins online database (http://string-db.org; version 12.0, Zurich, Switzerland), which functions as a retrieval tool for interacting genes, was utilized. Each node within the PPI network represents a specific gene, protein, or other biomolecule. The connections between these nodes signify the interactions that occur between these biomolecules. The nodes that are most closely connected may represent hub genes that possess significant physiological regulatory activities. Studying the functional connections between proteins might assist in understanding how diseases develop or spread, and interactions with scores >0.4 are regarded as statistically significant. Subsequently, we imported the STRING file into the Cytoscape software (version 3.9.1, Cytoscape Consortium, San Diego, California, USA) and proceeded to construct the PPI network diagram, which illustrated the interactions between the DEGs. Next, the gene file was imported into the Molecular Complex Detection plug-in (version 2.0.0, San Diego, California, USA) of Cytoscape software to screen the top 10 hub genes based on their degree value.
Patients
The subjects for immunohistochemical and immunocytochemical staining were selected from patients who met the criteria for low-risk PTMC, with the exception of LNMs and had undergone thyroid lobectomy and lymph node dissection. Owing to the low occurrence rate of lateral cervical LNM in PTMC, we excluded instances with such metastasis to concentrate on the research subjects. The following criteria were used to determine inclusion: (1) Adult patients who underwent lymph node dissection and lobectomy from January 1, 2018 to December 31, 2020; (2) Tumor’s maximum diameter was 1 cm or less, and there was neither extrathyroidal invasion nor distant metastasis; (3) The tumor was unifocal. The subsequent factors were considered exclusions: (1) Cases with incomplete clinical information; (2) PTMC that was multifocal; (3) PTMC that had metastasized to the lymph nodes in the lateral region of the neck; (4) PTMC variants that were classified as high-risk based on postoperative pathology results; (5) PTMC that had extrathyroidal invasion; and (6) PTMC with tracheal or recurrent laryngeal nerve invasion and distant metastasis. In general, except for the LNM status, all the cases screened by the inclusion and exclusion criteria possessed the characteristics of low-risk PTMC. These patients are the ones who have the greatest clinical need to predict cervical LNM.
The G-power software (version 3.1.9.7, Franz Faul, Kiel, Germany) was employed to ascertain the adequacy of our sample size. The power value was set at 0.9, which is a commonly accepted level for detecting meaningful effects. Through this analysis, it was determined that a sample size of 183 cases or more would be adequate to detect statistically significant differences in immunohistochemical and immunocytochemical staining.
The PTMCs that had been screened were divided into two groups based on their pathological lymph node status: the pN0 PTMC group, which had no pathological lymph node involvement, and the pN1a PTMC group, which had pathological central lymph node involvement. To attain a balanced representation of both groups in terms of tumor diameter, age, and sex, a 1:1 propensity score matching technique was applied with a caliper set to 0.2. The Scitb package (version 1.4.10, R Foundation for Statistical Computing, Vienna, Austria) within R software was utilized for this purpose.
Immunohistochemical and immunocytochemical staining
Reagents and antibodies
The primary antibodies employed were as follows: (1) Cytokeratin 5/6 (CK5/6, clone D5/16B4, dilution 1:50, Abcam, UK). (2) Chromogranin A (CgA, clone MX018, dilution 1:100, Maxim, China). (3) Pair box gene 2 (Pax-2, clone RMA-0816, dilution 1:100, Maxim, China). The secondary antibodies (Horseradish peroxidase goat anti-rabbit secondary antibody, item number ab6721, dilution 1:10000, Abcam, UK) and detection reagents were provided by the Ventana system and were selected based on their compatibility with the primary antibodies and the specific staining protocol requirements.
Tissue preparation and sample selection
(1) Surgical specimens: Four-micrometer-thick whole tissue sections of PTMC surgical specimens were acquired from patients who met the inclusion criteria described in the “Patients” section. These specimens were fixed in 10% formalin after resection and processed following standard histological techniques. Sections were cut from paraffin-embedded blocks and mounted on glass slides for immunohistochemical staining. (2) Preoperative thyroid FNA liquid-based preparations: Preoperative thyroid FNA liquid-based samples were collected from the same patients. The samples were prepared by means of a liquid-based cytology method to ensure optimal cell preservation and distribution.
Staining procedure
The immunohistochemical and immunocytochemical staining was conducted using the Ventana Benchmark Ultra immunostainer (Ventana Medical Systems, Tucson, Arizona, USA), which is a highly automated and reliable instrument for performing such assays. Heat-induced epitope retrieval was performed on the immunostainer using cell conditioning 1 buffer from Ventana Medical Systems. After blocking endogenous peroxidase activity, the slides were incubated with the following antibodies: for CK5/6, antigen retrieval was performed in citrate buffer (pH 6.0) for 20 min at 95°C; for CgA, in Ethylenediaminetetraacetic acid buffer (pH 8.0) for 15 min at 98°C; and for Pax-2, in citrate buffer (pH 6.0) for 18 min at 95°C. The signal was detected using a chromogenic substrate (3,3’-diaminobenzidine), and the reaction was developed for 5 min. The slides were then counterstained with hematoxylin for 1 min, dehydrated, cleared, and mounted with a coverslip.
Quality control and validation
CK5/6 expression was predominantly observed in the cytoplasm and cell membrane of tumor cells. CgA staining was mainly localized in the cytoplasm of tumor cells. Pax-2 expression was detected in the nucleus of tumor cells. The staining results were assessed independently by two experienced pathologists who were blinded to the clinical information of the patients. Any discrepancies in the interpretation were resolved through consensus discussion. The staining results were categorized as negative and positive.
Statistical analysis
A Chi-square test was employed to evaluate the disparity in immunohistochemical examination levels between the groups of PTMC patients with CLNM and those without metastasis using the statistical package for the social sciences (version 19.0, international business machines corporation, Armonk, NewYork, USA). If the P-value was >0.05, it was considered as statistical significance. For the chi-square test implementation, we selected between the Pearson Chi-square test and the continuity-corrected Chi-square test, taking into account both the total number of samples and the theoretical frequency. Additionally, we proceeded to calculate the effect size of our obtained result, which provides a more comprehensive understanding of the practical significance and magnitude of the associations detected in our study.
The pipeline of the “Materials and Methods” section is shown in Figure 1, which is created by Microsoft Office Word software (Microsoft Office 2017, Redmond, Washington, USA).
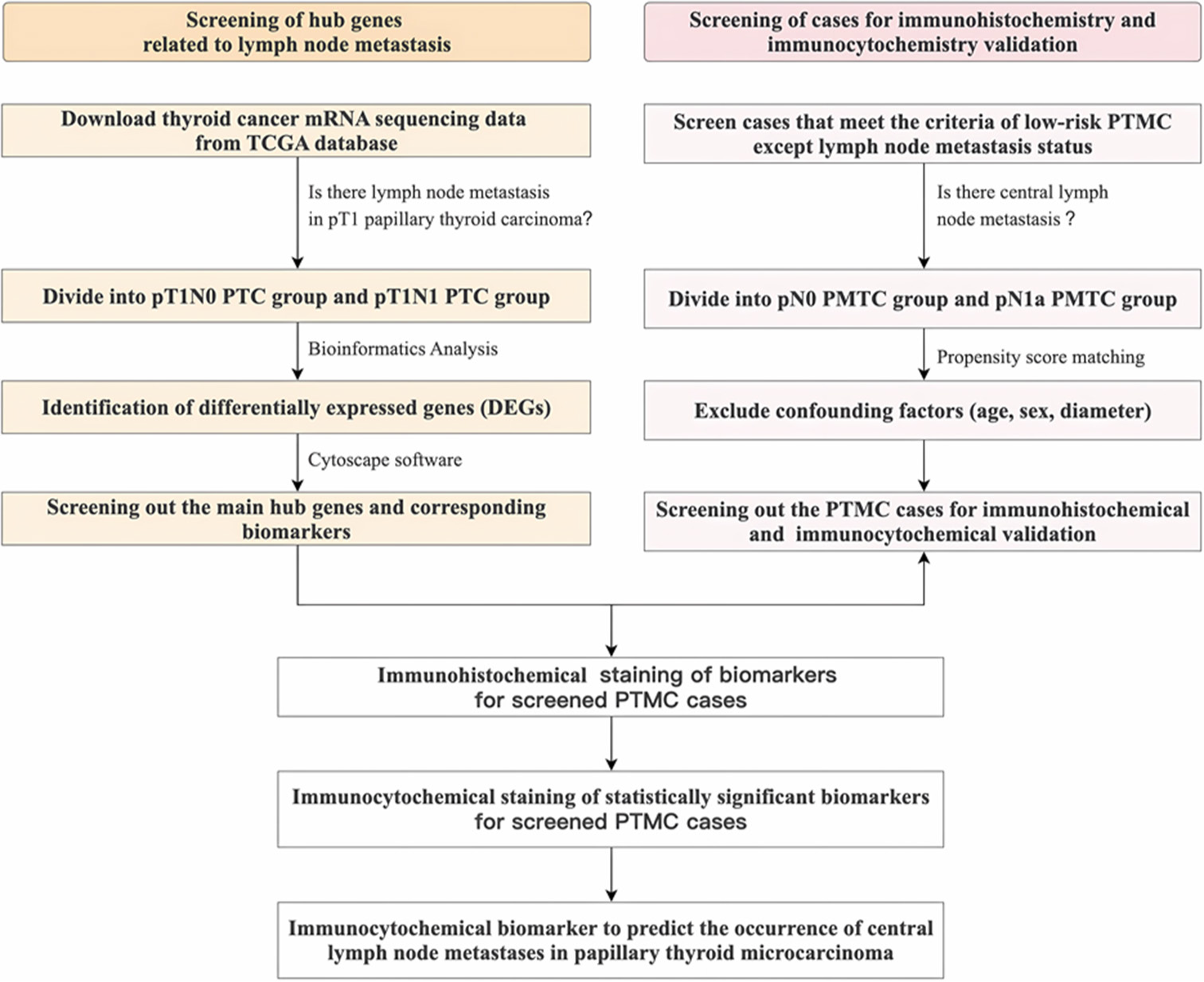
- The pipeline of the “Materials and Methods” section. mRNA: Messenger ribonucleic acid, TCGA: The cancer genome atlas, pT1: Pathological tumor stage 1, PTC: Papillary thyroid carcinoma, DEG: Differentially expressed gene, PTMC: Papillary thyroid microcarcinoma.
RESULTS
Hub genes related to LNM
A total of 132 instances of PTC with a pathological T stage of pT1 and a node stage were extracted from the TCGA database. This comprised 44 cases in the lymph node-positive group (pT1N1 PTC group) and 88 cases in the lymph node-negative group (pT1N0 PTC group). In accordance with the procedure described in the “Materials and Methods” section, a total of 292 DEGs were successfully identified. Among these, 87 genes were found to be up-regulated, while 205 genes were down-regulated. Figure 2 illustrates the heatmap and volcano plot of the DEGs.
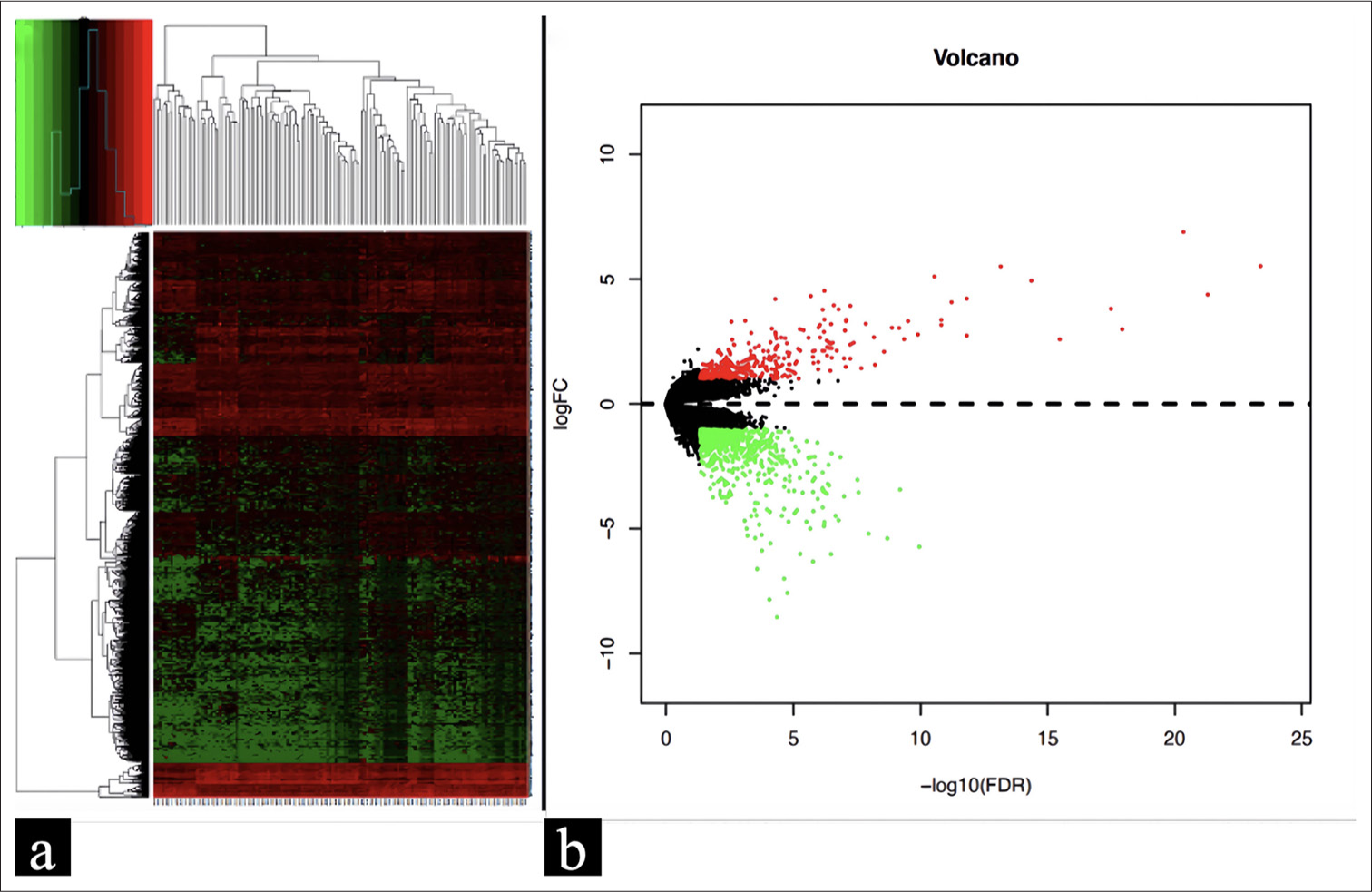
- (a) Heatmap of 292 DEGs retrieved from the TCGA database; (b) Volcano plot of 292 DEGs. The red color indicates the genes that have been upregulated, the green color represents the genes that have been downregulated, and the black color denotes genes without change. DEG: Differentially expressed gene, TCGA: The cancer genome atlas.
In addition, the STRING database was employed to construct a PPI network for the identified DEGs. Subsequently, the string file was imported into the Cytoscape software to screen for the top 10 hub genes. Figure 3 presents the top 10 hub genes with the highest degree value. Among these genes, KRT5, KRT6A, KRT14, KRT6B, and KRT16 are upregulated, whereas CHGA, Neuropeptide Y, Calcitonin Related Polypeptide Alpha, Parathyroid Hormone and PAX2 are downregulated.
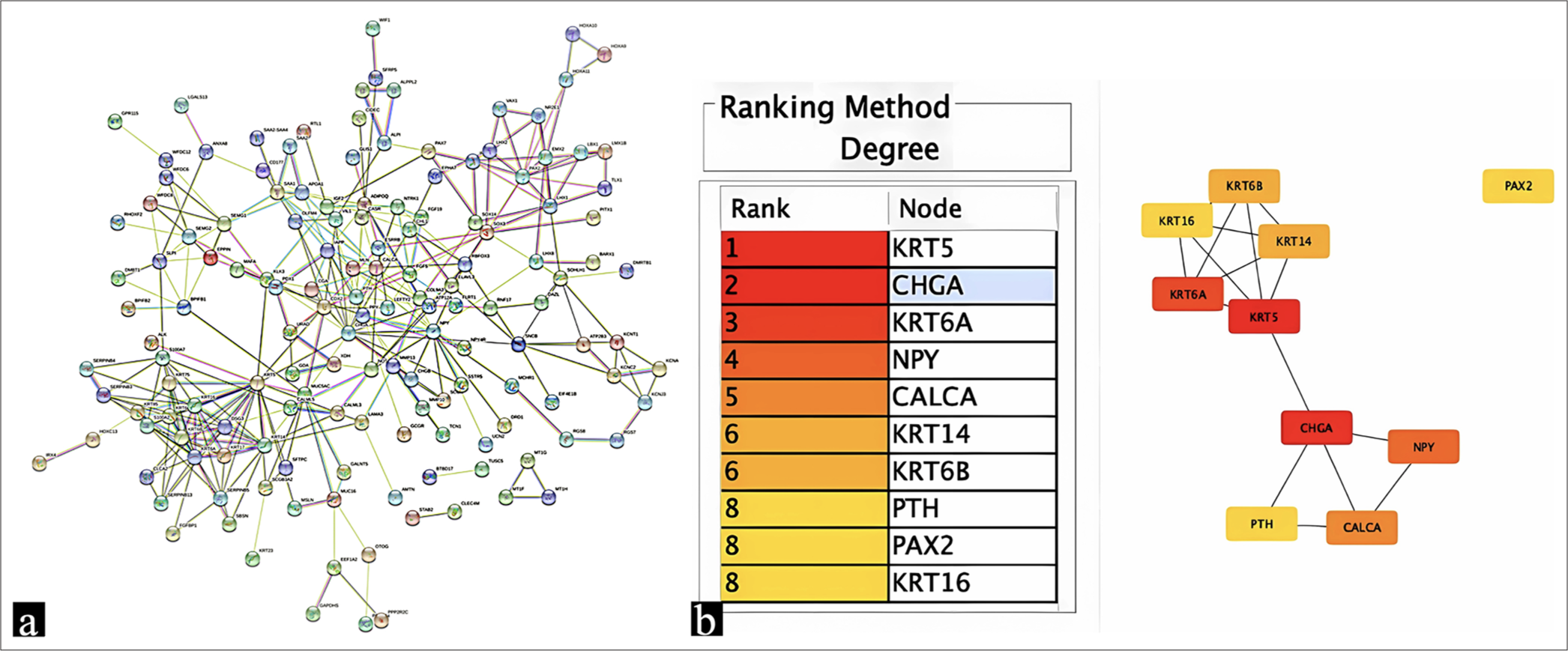
- (a) PPI network constructed among DEGs with the criterion of interaction score >0.4; (b) Top 10 hub genes in PPI relationship pairs were ranked in the table and depicted in the graph. PPI: Protein-protein interaction, DEG: Differentially expressed gene.
Immunohistochemical and immunocytochemical validation
The study screened 272 suitable cases out of a total of 2091 instances of PTC based on the criteria for inclusion and exclusion. After using propensity score matching to ensure equivalence in age, gender, and diameter between the two groups, a total of 104 cases with CLNM and 104 instances without LNM were selected for further immunohistochemical staining [Figure 4a, created by Microsoft Office Word software]. Propensity score matching demonstrated that the standardized mean difference (SMD) after matching was below 0.1 [Figure 4b]. Typically, an SMD <0.1 implies a favorable balance when assessing group differences, indicating that the disparity between the study groups is minimal.
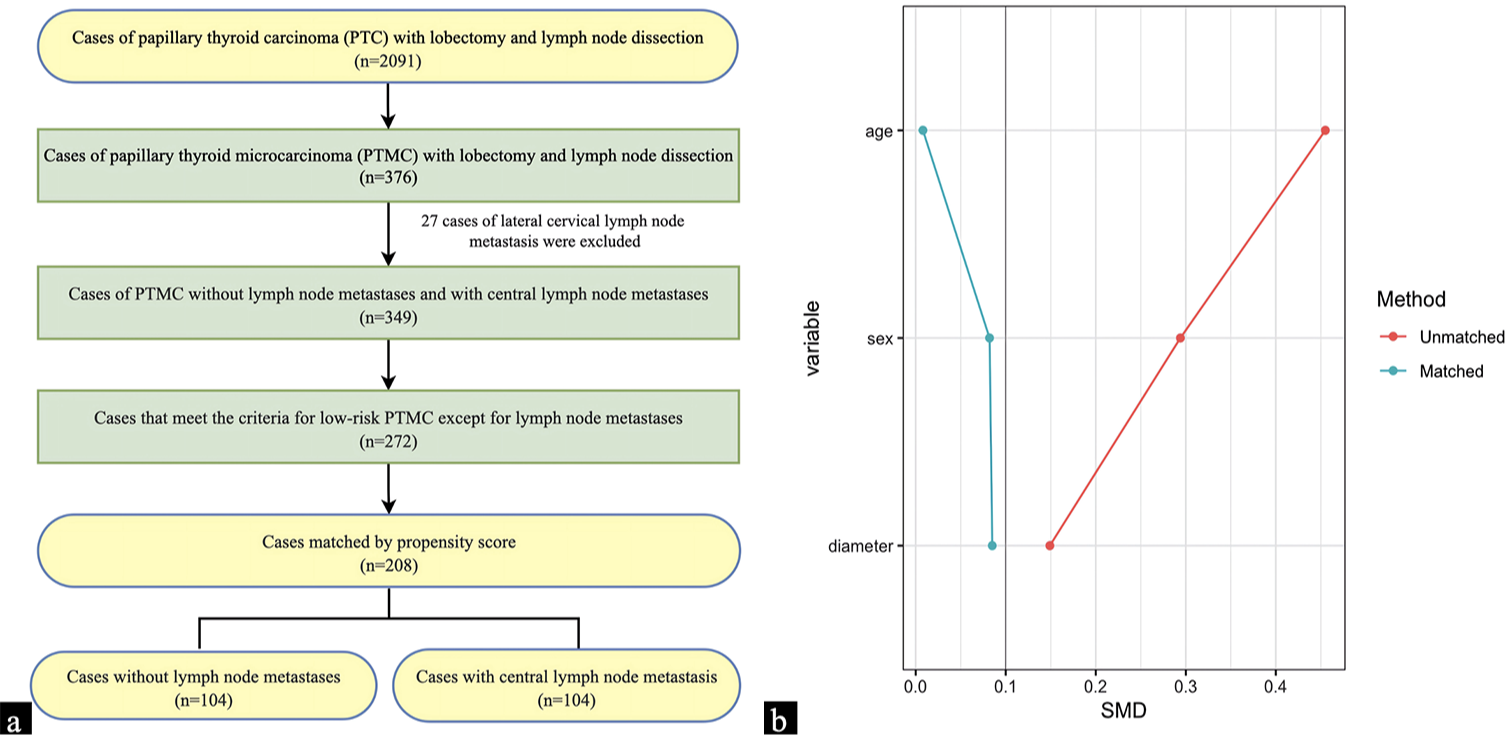
- (a) The screening process of cases for immunohistochemical and immunocytochemical validation; (b) The standardized mean difference between the two groups of cases before and after propensity score matching.
Among the 208 cases, the biomarkers (CK5/6, CgA, and Pax-2) corresponding to the main hub genes (KRT5, KRT6A, CHGA, and PAX2) were immunohistochemically stained in postoperative thyroidectomy specimens. The immunohistochemical staining results revealed a statistically significant difference in the expression of CK5/6 between pN1a PTMC group and pN0 PTMC group (P = 0.002). PTMC cases with CLNM were more likely to be positive for CK5/6, the positive model mainly displayed spot-positive characteristics at the periphery of the tumor lesion, with a small part exhibiting strong patchy positivity [Figure 5a-d]. However, the expression levels of CgA (P = 0.208) and Pax-2 (P = 0.999) did not show any significant differences between the two groups [Table 1].
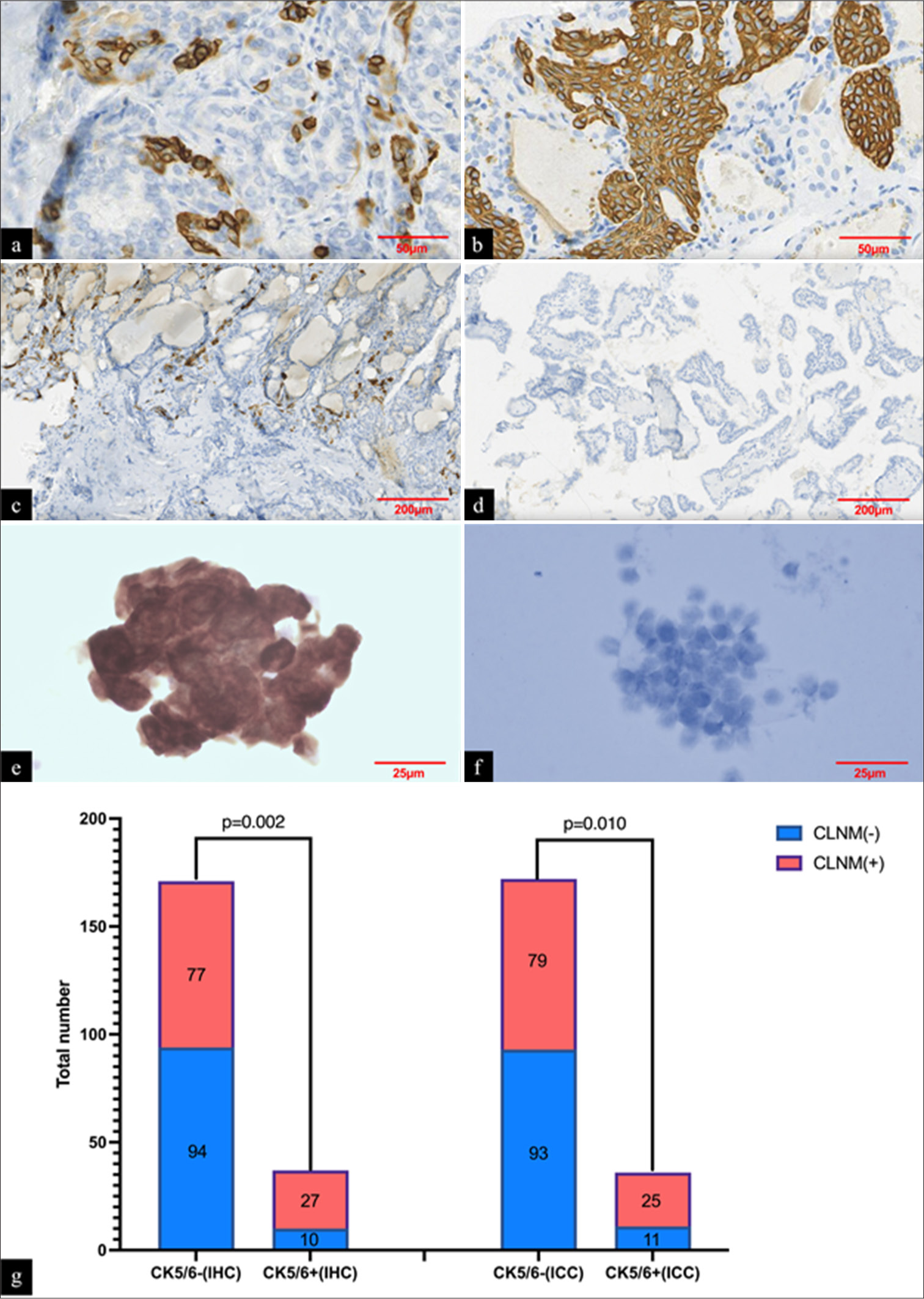
- (a) PTMCs with lymph node metastases were more likely to display spot-positive CK5/6 expression (immunohistochemical staining, ×400); (b) A small number of PTMCs with lymph node metastases were intensely patchy-positive for CK5/6 (immunohistochemical staining, ×400); (c) CK5/6 positive sites with lymph node metastases were usually predominantly located at the periphery of the tumor lesion (immunohistochemical staining, ×100). (d) CK5/6 negative staining result (immunohistochemical staining, ×100). (e and f) Presentation of positive and negative results of CK5/6 immunocytochemical staining conducted in preoperative thyroid FNA liquid-based samples (immunocytochemical staining, ×800). (g) The staining results of CK5/6 in immunohistochemistry and immunocytochemistry. PTMC: Papillary thyroid microcarcinoma, CK5/6: Cytokeratin 5/6, FNA: Fine needle aspiration, IHC: Immunohistochemistry, ICC: Immunocytochemistry, CLNM: Central lymph node metastasis.
Subsequently, CK5/6 immunocytochemical staining was conducted in preoperative thyroid FNA liquid-based samples, and the results further verified that CK5/6 was more likely to be positively expressed in PTMCs with CLNM (P = 0.010) [Figure 5e-g].
| Biomarkers | CLNM | P | Effect size (Phi coefficient) | Odds ratio (95% confidence interval) | |
|---|---|---|---|---|---|
| − | + | ||||
| CK5/6 | 0.002 | 0.214 | 3.296 (1.489–6.951) | ||
| − | 94 | 77 | |||
| + | 10 | 27 | |||
| CgA | 0.208 | 0.087 | 1.709 (0.7673–3.920) | ||
| − | 94 | 88 | |||
| + | 10 | 16 | |||
| Pax-2 | 0.999 | 0.070 | +infinity (0.1111 –+infinity) | ||
| − | 104 | 103 | |||
| + | 0 | 1 | |||
CLNM: Central lymph node metastasis, CK5/6: Cytokeratin 5/6, CgA: Chromogranin A, Pax-2: Pair box gene 2
DISCUSSION
Numerous studies have documented molecular markers correlated with the onset and progression of PTC.[11] Ruiz and colleagues performed an analysis of DEGs related to LNM in PTC and identified a panel of 25 genes that can be utilized to differentiate between the pN0 and pN1 groups of PTC samples. These genes exhibited a sensitivity of 86% for detecting LNM.[12] Zhang et al. reported that an 11-gene signature displayed good discriminative performance for lymph node metastasis (LNM) prediction (area under the curve [AUC] = 0.847), and TIPARP mRNA was upregulated in PTC with LNM versus with-out (AUC = 0.692).[13] Moreover, Napoli et al. identified that four micro ribonucleic acids (miR-154-3p, miR-299-5p, miR-376a-3p, and miR-302E) exhibited a significant differential expression in cytological samples of PTC with LNM and predicted the positive nodal status with relatively good performance (AUC = 0.66–0.72).[14] In general, a combination of DEGs is capable of determining whether PTC has LNM. This method demonstrates reliability and scientific rigor, making it a valuable tool for predicting PTMC LNM. Nevertheless, the genes implicated in LNM in PTC may vary depending on the tumor diameter. Hence, it is crucial to investigate the specific genes associated with LNM in PTMC. Li et al. identified 20 DEGs in the group of patients with LNMs (N1) and those without (N0) in PTMC. Among these DEGs, FN1 demonstrated good performance in differentiating between N1 and N0 patients, with an AUC value of 0.690.[15] The study carried out by Lin et al. revealed that interferon-stimulated gene 15 was highly expressed in PTMC with LNM (P < 0.001).[16] However, these studies either exhibited insufficient prediction accuracy or had a limited validation of cases, and have not explicitly focused on low-risk PTMC, which holds the utmost therapeutic significance.
The current study explored the genes that affect LNM through the application of bioinformatics analysis, immunohistochemistry, and immunocytochemistry. Overall, a comprehensive analysis identified a total of 292 DEGs, and the top 10 hub genes were uncovered from a dataset of 122 instances of pT1 PTCs. Further immunohistochemical and immunocytochemical validation of the primary hub genes demonstrated notable disparities in CK5/6 between the PTMC group with CLNM and those without LNM. During this analytical process, the Chi-square test was adopted to evaluate the disparities in immunohistochemical examination levels between PTMC patients with and without CLNM. When conducting the test, either the Pearson Chi-square test or the continuity-corrected Chi-square test was selected based on the total number of samples and the theoretical frequency, thereby revealing the differences among the groups. This study fully acknowledges the potential interference of confounding factors on CK5/6 expression and CLNM development. Therefore, in terms of patient characteristics, factors such as age, gender, and tumor size that might affect the research results were taken into account. During the sample screening stage, the propensity score matching method was utilized to accurately match PTMC patients with and without CLNM according to age, gender, and tumor diameter. The SMD between groups was stringently controlled below 0.1, effectively diminishing the confounding effects of these factors and enhancing the comparability between groups. This guarantees that the differences in CK5/6 expression are attributed to the CLNM status rather than the disparities in patients’ basic characteristics, all of which have laid a relatively solid statistical foundation for the research conclusions. It is noteworthy that the cases utilized for verification were those that fulfilled the criteria of low-risk PTMC, with the exception of the status regarding LNM. From a therapeutic perspective, the selection of these cases is of utmost clinical significance, as the presence or absence of LNM in these cases will lead to significantly different treatment strategies.
CK5/6 is a frequently utilized antibody in the field of clinical pathology. It is predominantly detected in squamous epithelium, ductal epithelial basal cells, myoepithelial cells, and mesothelial cells in normal tissues. In thyroid cancer, CK5/6 can be employed to assist in the diagnosis of thyroid squamous cell carcinoma and can also be used to diagnose the squamous cell component in PTC, although the latter is less used in clinical practice due to the paucity of literature documenting the role of the squamous component in PTC.
Squamous cell carcinoma of the thyroid is postulated to have originated from PTC, the former is associated with a poorer prognosis and a higher incidence of LNMs.[1,17] Consequently, the presence of a squamous cell component in PTC may imply an elevated likelihood of LNM, a hypothesis that is further supported by some studies.[18] Interestingly, there are reports suggesting that in basal-like breast cancer cell lines, CK5 knockdown resulted in a reduction in LNMs in vivo compared to control cells,[19] although this was not directly observed in PTC. In the context of thyroid cancer, despite the dearth of direct evidence, it is plausible that CK5/6 could be implicated in processes such as epithelial-mesenchymal transition (EMT).[19] EMT is a crucial process in cancer metastasis, during which epithelial cells acquire mesenchymal traits, endowing them with the ability to migrate and invade neighboring tissues. CK5/6 may potentially contribute to maintaining the epithelial phenotype or modulating EMT-related signaling pathways, thereby exerting an influence on the metastatic potential of PTMC cells. Moreover, CK5/6 has been proposed as a possible stem cell marker.[20] Stem cells are known to interact with other molecules or signaling cascades involved in lymphangiogenesis,[21] the formation of new lymphatic vessels that facilitate the spread of cancer cells to regional lymph nodes. These investigations and hypotheses have, to some extent, demonstrated that CK5/6 is capable of predicting LNM in PTMC, thereby validating the reliability of our research findings.
The novelties of this study are summarized as follows: (1) This study represents the first instance of using commonly employed antibodies in clinical practice to predict the presence of CLNM in PTMC. CK5/6 is a widely utilized antibody in daily pathology work. The findings of this study can be promptly translated into clinical practice to facilitate the prediction of the status of PTMC central lymph nodes. (2) For cases that underwent immunohistochemical and immunocytochemical verification, we employed strict inclusion criteria and propensity score matching to select 208 PTMC cases from 2091 cases. These cases fulfilled the criteria for low-risk PTMC except for LNM.
This study exhibits the following limitations: (1) This study is a single-center study, and multicenter investigations are warranted for further validation. (2) The bioinformatics analysis was conducted using mRNA sequencing data of pT1 PTCs (≤2 cm), while immunohistochemical and immunocytochemical staining validation was performed on specimens of PTMC patients (≤1 cm). Owing to the absence of maximum diameter information about the tumor in the TCGA database, we could only select pT1 PTC in the bioinformatics analysis. In immunohistochemical and immunocytochemical verification, we selected PTMC, which is of greater clinical relevance. (3) There may be other confounding factors that could have affected CK5/6 expression or CLNM development. For example, patient-related factors were not thoroughly examined in relation to CK5/6 expression. Moreover, the immunohistochemical and immunocytochemical staining methods possess inherent limitations, such as inter-observer variability in the assessment of staining intensity and percentage of positive cells. Despite using two experienced pathologists and a standardized scoring system, a certain level of subjectivity may still exist. (4) The study did not investigate the functional consequences of CK5/6 expression in PTMC cells, which could yield further insights into its role in LNM.
Looking ahead to future research directions in the field of clinical medicine, actively promoting multi-center and prospective studies holds profound significance. By extensively incorporating samples from diverse regions and various clinical backgrounds, the universality and stability of the conclusions drawn from this study can be comprehensively verified across a wide range of medical settings. This will facilitate an in-depth analysis of the precise efficacy and long-term prognostic value of CK5/6 as a biomarker in clinical decision-making, thereby laying a solid empirical foundation for formulating unified and efficient clinical guidelines. In the realm of basic medicine, focusing on CK5/6 gene knockout and rigorous validation at the cell line level is of crucial importance. Through the application of precise gene editing techniques to construct knockout models, an in-depth exploration into the impact mechanisms of CK5/6 gene deletion on key tumor biological behaviors such as cell proliferation, migration, invasion, and EMT can be conducted. This will elucidate its functional role in the lymphatic metastasis process of papillary thyroid microcarcinoma from the molecular origin, providing robust support for macroscopic clinical findings through microscopic mechanism analysis at the cellular level. Thus, a closely intertwined and synergistic research framework between basic and clinical research will be formed, opening up new avenues and injecting innovative impetus into the conquest of lymphatic metastasis in thyroid cancer.
The findings of this study harbor potential clinical implications. CK5/6 could be integrated into existing diagnostic protocols for PTMC. For instance, in cN0 PTMC patients, CK5/6 immunohistochemical staining of preoperative thyroid FNA samples could be performed. A positive CK5/6 expression might initiate further evaluation, such as a more detailed preoperative imaging study, or entail consideration of a more aggressive treatment option, such as prophylactic central lymph node dissection.
In comparison to established markers like BRAF or RET/PTC mutations linked to CLNM in thyroid cancer, CK5/6 offers a unique perspective. The combination of CK5/6 analysis with these genetic markers may provide a more comprehensive evaluation of the CLNM risk in thyroid cancer patients. When combined with other biomarkers, CK5/6 can augment the accuracy of CLNM prediction. Future studies could investigate the development of a biomarker panel comprising CK5/6 and other relevant markers to enhance PTMC patient risk stratification.
SUMMARY
This study employed bioinformatics analysis to detect hub genes and subsequently performed immunohistochemical and immunocytochemical staining of these genes in PTMC. CK5/6 has been established as a potential immunohistochemical and immunocytochemical biomarker for distinguishing PTMC with CLNM from PTMC without LNM. Future research with larger scale, prospective design, and in-depth exploration at the cell line level will contribute to further enhancing the credibility of research conclusions.
AVAILABILITY OF DATA AND MATERIALS
The data analyzed in this study can be accessed through the corresponding author upon reasonable request.
ABBREVIATIONS
CgA: Chromogranin A
CK5/6: Cytokeratin 5/
CLNM: Central lymph node metastasis
DEG: Differentially expressed gene
FDR: False discovery rate
FNA: Fine needle aspiration
ICC: Immunocytochemistry
IHC: Immunohistochemistry
LNM: Lymph node metastasis
mRNA: Messenger ribonucleic acid
Pax-2: Pair box gene 2
PPI: Protein-protein interaction
pT1: Pathological tumor stage 1
PTC: Papillary thyroid carcinoma
PTMC: Papillary thyroid microcarcinoma
SMD: Standardized mean difference
STRING: Search tool for the retrieval of interacting genes
TCGA: The cancer genome atlas
ACKNOWLEDGMENT
We acknowledge the contributions of the TCGA program registries for creating and updating the TCGA database.
AUTHOR CONTRIBUTIONS
LLR: Study design, data collection and analysis, and manuscript drafting; JW, QW and YLZ: Data collection, cytology review, and statistical analysis; WHR: Study design, data collection and analysis, and manuscript review. All authors have read and approved the final version of the manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of the Peking University Cancer Hospital (approval ID: No. 2023KT72). Written informed consent was obtained from all the patients or their families prior to the enrollment of this study. The study was conducted in accordance with the Declaration of Helsinki.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: This study was funded by Science Foundation of Peking University Cancer Hospital, Grant/Award Number: PY202302.
References
- Overview of the 2022 WHO classification of thyroid neoplasms. Endocr Pathol. 2022;33:27-63.
- [CrossRef] [PubMed] [Google Scholar]
- Long-term outcomes and risk factors of radiofrequency ablation for T1N0M0 papillary thyroid carcinoma. JAMA Surg. 2024;159:51-8.
- [CrossRef] [PubMed] [Google Scholar]
- Indications and strategy for active surveillance of adult low-risk papillary thyroid microcarcinoma: Consensus statements from the Japan association of endocrine surgery task force on management for papillary thyroid microcarcinoma. Thyroid. 2021;31:183-92.
- [CrossRef] [PubMed] [Google Scholar]
- Active surveillance for low-risk thyroid cancers: A review of current practice guidelines. Endocrinol Metab (Seoul). 2024;39:47-60.
- [CrossRef] [PubMed] [Google Scholar]
- Surgical treatment in papillary thyroid microcarcinoma. Sisli Etfal Hastan Tip Bul. 2018;52:244-8.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnostic value of contrast-enhanced ultrasonography in the preoperative evaluation of lymph node metastasis in papillary thyroid carcinoma: A single-center retrospective study. BMC Surg. 2023;23:325.
- [CrossRef] [PubMed] [Google Scholar]
- Exploring risk factors for cervical lymph node metastasis in papillary thyroid microcarcinoma: Construction of a novel population-based predictive model. BMC Endocr Disord. 2022;22:269.
- [CrossRef] [PubMed] [Google Scholar]
- Clinicopathologic factors and preoperative ultrasonographic characteristics for predicting central lymph node metastasis in papillary thyroid microcarcinoma: A single center retrospective study. Braz J Otorhinolaryngol. 2022;88:36-45.
- [CrossRef] [PubMed] [Google Scholar]
- A novel tool for predicting the risk of central lymph node metastasis in patients with papillary thyroid microcarcinoma: A retrospective cohort study. BMC Cancer. 2022;22:606.
- [CrossRef] [PubMed] [Google Scholar]
- differential diagnosis and prognostic significance of squamous cell components in papillary thyroid carcinoma. Chin J Clin Exp Pathol. 2023;9:1482-6.
- [Google Scholar]
- Clinical application of next-generation sequencing in advanced thyroid cancers. Thyroid. 2022;32:657-66.
- [CrossRef] [PubMed] [Google Scholar]
- A novel gene panel for prediction of lymph-node metastasis and recurrence in patients with thyroid cancer. Surgery. 2020;167:73-9.
- [CrossRef] [PubMed] [Google Scholar]
- TIPARP as a prognostic biomarker and potential immunotherapeutic target in male papillary thyroid carcinoma. Cancer Cell Int. 2024;24:34.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA profiling predicts positive nodal status in papillary thyroid carcinoma in the preoperative setting. Cancer Cytopathol. 2022;130:695-704.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of potential diagnostic and prognostic biomarkers for papillary thyroid microcarcinoma (PTMC) based on TMT-labeled LC-MS/MS and machine learning. J Endocrinol Invest. 2023;46:1131-43.
- [CrossRef] [PubMed] [Google Scholar]
- Deciphering novel biomarkers of lymph node metastasis of thyroid papillary microcarcinoma using proteomic analysis of ultrasound-guided fine-needle aspiration biopsy samples. J Proteomics. 2019;204:103414.
- [CrossRef] [PubMed] [Google Scholar]
- Conditional survival analysis between primary squamous cell carcinoma of the thyroid and anaplastic thyroid carcinoma. Int J Surg. 2024;111:1558-60.
- [CrossRef] [PubMed] [Google Scholar]
- Squamous differentiation in papillary thyroid carcinoma: A rare feature of aggressive disease. J Surg Res. 2018;223:39-45.
- [CrossRef] [PubMed] [Google Scholar]
- Cytokeratins 5 and 17 maintain an aggressive epithelial state in basal-like breast cancer. Mol Cancer Res. 2022;20:1443-55.
- [CrossRef] [PubMed] [Google Scholar]
- The breast stem cell (CK5/6(+)) concept and its relation to the diagnosis of benign and malignant ductal epithelial hyperplasia. Zhonghua Bing Li Xue Za Zhi. 2013;42:73-7.
- [Google Scholar]
- Tube-like structures with co-expression of D2-40 and CD34: Newly formed vasculatures? Int J Biol Sci. 2012;8:1206-16.
- [CrossRef] [PubMed] [Google Scholar]








