Translate this page into:
KRAS detection on archival cytological smears by the novel fully automated polymerase chain reaction-based Idylla mutation test
*Corresponding author
-
Received: ,
Accepted: ,
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background:
Molecular techniques are relevant to modern cytopathology, but their implementation is difficult without molecular expertise and infrastructure. The assessment of KRAS mutational status on cytological preparations may be useful either to refine uncertain diagnoses on pancreatic aspirates or to yield predictive information to plan targeted treatment of metastatic colorectal cancer (mCRC). The novel test Idylla™ enables fully automated KRAS genotyping in approximately 2 h, even in less experienced hands.
Materials and Methods:
This study aims to validate this methodology to detect KRAS mutations on archival cytological preparations of pancreatic cancer (n = 9) and mCRC (n = 9) by comparing the Idylla™ performance to that of standard real-time polymerase chain reaction.
Results:
The same 11 mutations (n = 4: p.G12D; n = 2: p.G12V; n = 2: p.A59E/G/T; n = 1: p.G12R; n = 1: p.G13D; n = 1: p.Q61H) were detected by both techniques.
Conclusion:
Even in less experienced laboratories, a cytopathologist may easily integrate morphological diagnostic report with accurate KRAS mutation detection, which is relevant for diagnostic and treatment decisions.
Keywords
Aspiration cytology
Idylla
KRAS
molecular pathology
INTRODUCTION
Molecular techniques are more and more significant in daily cytology practice.[1] In particular, the demonstration of specific oncogenic mutations is a relevant diagnostic and predictive tool.[1] As an example, the assessment of KRAS mutational status may be useful to refine uncertain cytological diagnoses of solid pancreatic lesions[2] and to differentiate mucinous from nonmucinous cysts.[3] Moreover, metastatic colorectal cancer (mCRC) patients harboring KRAS mutations at codons 12 and 13 in exon 2, codons 59 and 61 in exon 3, and codons 117 and 146 of exon 4 do not benefit from the administration of EGFR antagonists.[4] In routine practice, KRAS testing is usually performed on resected primary tumor samples of CRC, but the cytological sampling of the metastatic sites is useful when there is complete response to neoadjuvant chemoradiotherapy in patients with rectal cancer[5] and to serially monitor for the emergence of mutant treatment resistant clones.[67]
In both pancreatic and colorectal cancer patients, KRAS testing should have a fast turnaround testing, in line with the need of urgent clinical actions. Thus, instead of outsourcing cytological samples to a small number of referral molecular pathology laboratories, KRAS testing may be carried out in the same center where the patient is diagnosed. Unfortunately, the competence needed to validate and run complex molecular diagnostic assays is beyond most cytopathology laboratories.[8] However, the automated allele-specific real-time polymerase chain reaction (RT-PCR) technology is advancing at rapid pace. In particular, the fully automated molecular diagnostics system Idylla™ (Biocartis, Mechelen, Belgium) is a fascinating technology.[91011] Sample preparation is combined with PCR thermocycling and fluorescence detection of target sequences. Without needing highly skilled staff, within approximately 90 min, the Idylla™ mutational tests approved for in vitro diagnostic use by the European Community (CE-IVD) marked can genotype KRAS with 5% detection limit, enabling method standardization even in those diagnostic units without molecular infrastructures.[12] Although the Idylla™ tests were designed for use with formalin-fixed paraffin-embedded (FFPE) sections, as we previously have shown the Idylla™ system can also process DNA preparations obtained from fresh cells.[12] The aim of this study was to assess Idylla™ performance on the DNA extracted from archival smears of pancreatic and mCRC.
MATERIALS AND METHODS
Study cases
The database of the Cytopathology Department at the University of Naples was searched to select archival smears of pancreatic cancer (n = 10) and metastatic adenocarcinoma of colorectal origin (n = 11). The smears were reviewed by two qualified pathologists, EV and MdA, and the diagnosis was confirmed, and in all cases, the purity of tumor cells was estimated as a percentage of malignant cells out of the total nucleated cells. Since the analytic sensitivity of the Idylla test is high (5% of mutant allele detection), cases were selected without a specific requirement of neoplastic cellularity. Archival smears were processed between March and May 2016. A single slide was retrieved for each case. Each slide was incubated in xylene for 3 days to allow coverslip removing and air-dried; tumor cells were scraped off directly from the whole glass surface. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Crawley, West Sussex, UK) and quantified (ng/µl) as previously described.[13] All cases were tested by Idylla regardless from the DNA concentration.
Since KRAS mutational analysis is part of the routine diagnostic workup of patients with pancreatic and colorectal lesions, the need for Ethic Committee's approval was not necessary for this study, in accordance with Medical Ethical Guidelines of the University Federico II Medical School. Accordingly to these guidelines, a comprehensive written informed consent was signed for the procedure (fine needle aspiration) that produced the tissue samples. All information regarding the human material was managed using anonymous numerical codes. All samples were handled in compliance with the Declaration of Helsinki (http://www.wma.net/en/30publications/10policies/b3/).
The Idylla™ system
The Idylla system is an automated platform for the detection of genetic mutations based on qualitative allele-specific RT-PCR.[91011] The apparatus features a sample preparation module integrated with a combined PCR thermocycling and fluorescence detection module. All consumables required to perform sample preparation and RT-PCR detection are provided in disposable cartridges that are loaded onto the Idylla system to enable the simultaneous detection of up to 30 molecular targets. Closing of the cartridge after inserting the sample avoids cross contamination.
The Idylla™ KRAS mutation test
The Idylla™ KRAS mutation test is a single-use cartridge-based test. Through microfluidic channels in the cartridge, nucleic acids are transported into five separate PCR chambers, which contain predeposited PCR reagents in dried form (i.e., primers, probes, enzymes) designed for the qualitative detection of 21 KRAS mutations and including KRAS total (wild-type gene acting as sample processing control).
Seven mutations are targeted in exon 2: p.G12C (c.34G>T), p.G12R (c.34G>C), p.G12S (c.34G>A), p.G12A (c.35G>C), p.G12D (c.35G>A), p.G12V (c.35G>T), and p.G13D (c.38G>A). Nine mutations can be detected in exon 3: p.A59E (c.176C>A), p.A59G (c.176C>G), p.A59T (c.175G>A), p.Q61K (c.181C>A; c.180_181TC>AA), p.Q61 L (c.182A>T), p.Q61R (c.182A>G), and p.Q61H (c.183A>C; c.183A>T); whereas five mutations are targeted in exon 4: p.K117N (c.351A>C; c.351A>T), p.A146P (c.436G>C), p.A146T (c.436G>A), and p.A146V (c.437C>T).
To perform the analysis, 10 ng of extracted genomic DNA was directly pipetted inside an Idylla KRAS mutation test cartridge. This latter was, then, loaded into the Idylla instrument. The fully automated analysis required approximately 2 h. The Idylla console autoanalyzed the PCR curve to determine the presence or absence of a KRAS mutation and the results were presented on screen as either “no mutation detected” or “KRAS mutation detected,” indicating the specific mutation.
To assess the Idylla KRAS mutation test performance, in any single case, extracted DNA was run in parallel by the CE-IVD marked Easy KRAS kit (Diatech Pharmacogenetics, Jesi, Italy) on a QuantStudio 5 instrument (Thermo Fisher, Monza, Italy).[14] This commercially available test detects all relevant mutations of exon 2, 3, and 4 of KRAS by standard RT-PCR.
RESULTS
Idylla yielded valid results in 18/21 (85.7%) samples [Table 1] in a first run. Three samples, whose DNA concentration was <0.5 ng/µl, gave an invalid result. Only one of these cases was successfully amplified by the Easy KRAS kit, resulting wild type. On the overall, the group of 18 cases, was equally composed (n = 9) by pancreatic and mCRC smears. Papanicolaou-stained smears were more frequent (n = 11) than Diff-Quik-stained smears (n = 7). A total of 11 cases showed a KRAS mutation, and a representative example is reported in Figure 1. In all cases, the Easy KRAS kit confirmed the obtained results [Table 1]. In particular, the mutations detected by both techniques were the following: p.G12D (n = 4), p.G12V (n = 2), p.G12R (n = 1), p.G13D (n = 1), p.A59E/G/T (n = 2), p.Q61H (n = 1). Four mutant cases (p.G12D, p.G12R, p.G12V, and p.Q61H) were observed in the group of pancreatic smears, whereas seven mCRC smears harbored a KRAS mutation (n = 3 p.G12D; n = 2 p.A59E/G/T; n = 1 p.G12V; n = 1 p.G13D). The absence of mutations in five pancreatic and two mCRC smears was also confirmed by Easy KRAS kit. Noteworthy, most of the wild-type pancreatic cancer featured a low cellularity with the presence of <20% of cancer cells.
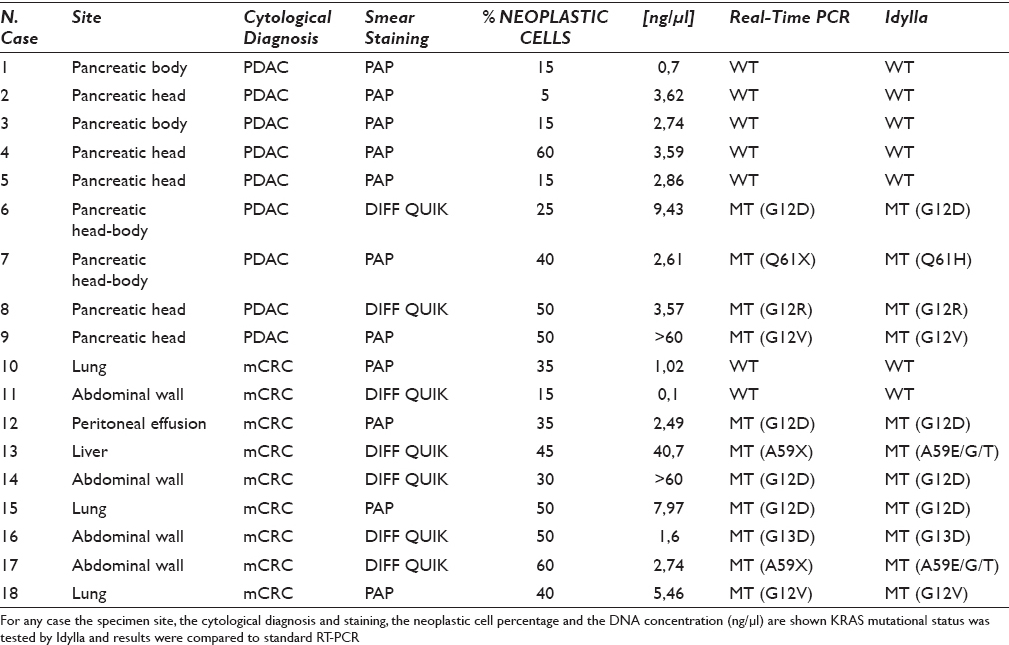
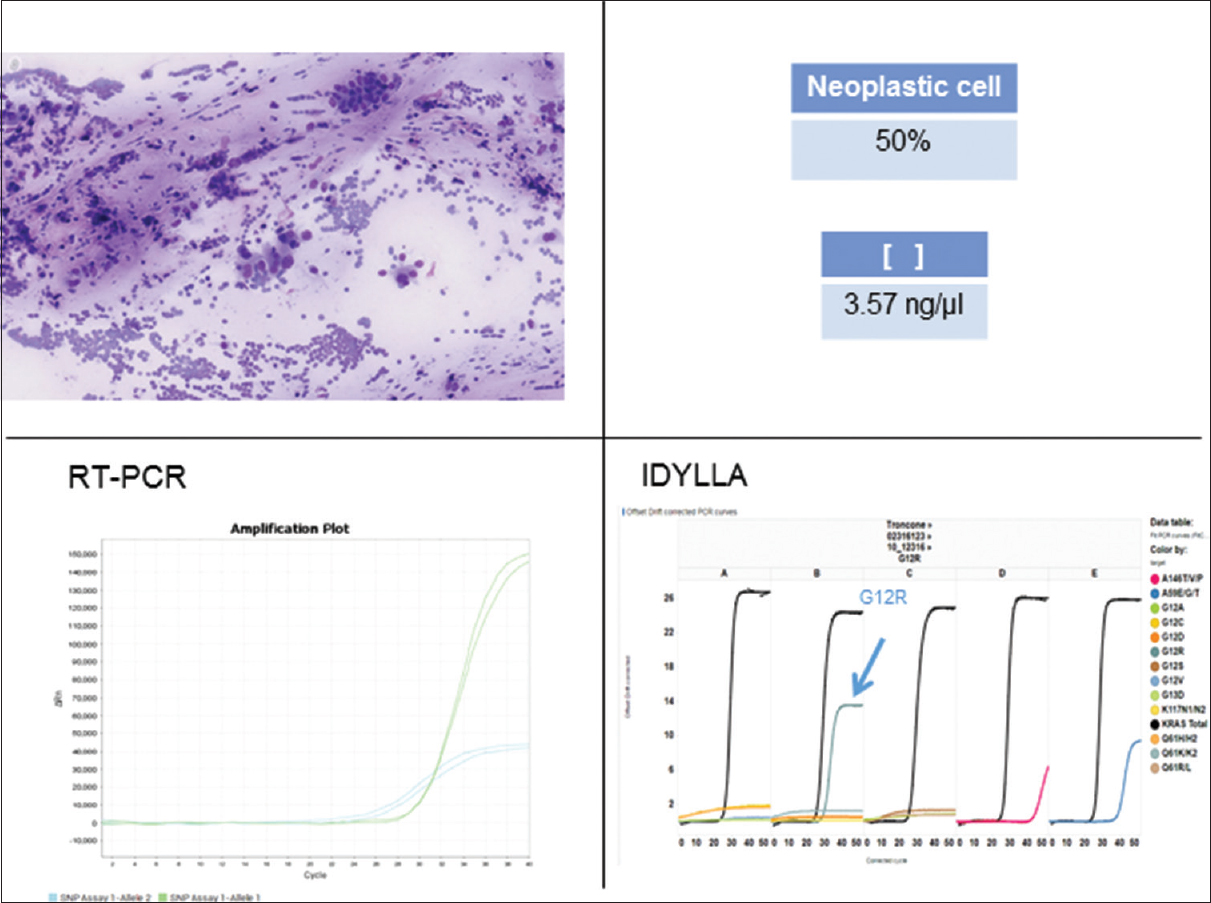
- Example of concordant molecular results in a KRAS exon 2 mutated sample (case #8, Table 1). Archival DNA was extracted from a Diff-Quik-stained smear of pancreatic cancer featuring 50% of neoplastic cells and a concentration of 3, 57 ng/μl. Representative graphs of standard real-time polymerase chain reaction and Idylla are reported both showing G12R KRAS mutation
DISCUSSION
On pancreatic fresh cells, collected by fine-needle aspiration and directly immersed in a tube with a preserving buffer, Idylla™ KRAS test showed a clinical sensitivity higher than Sanger sequencing.[12] Similar to that observed on pancreatic fresh cells, also on archival smears, the Idylla™ KRAS test performance is high and similar to that of Easy KRAS kit. In fact, the two methods showed a complete concordance, alleviating the concern of possible false negative results by Idylla. According to manufacturer, the minimum tumor percentage to avoid false negative results is 10%. Thus, there was only one case (#2) with really low tumor (5%) that does not rule out the possibility of false negative results by Idylla. However, recent cell line dilution data showed that the EGFR L858R point mutation can be detected by Idylla even at 1% dilution.[15]
Noteworthy, the rate of wild-type pancreatic adenocarcinoma was higher than the documented rates in most other published series.[1] This may reflect the limited number of cases tested but may also be explained by the low (<20%) neoplastic cellular content of most (4/5) cases. In addition, due to the stochastic distribution of benign and malignant cells, in these cases, manual microdissection to enrich for neoplastic cells was unfeasible, leading to a further dilution of potentially present mutant alleles. These limitations may be overcome by the more sensitive next-generation sequencing,[16] whose implementation is cost-effective only in large volume centralized laboratories.[17] Conversely, we showed that Idylla may be easily adopted by a large number of cytopathology laboratories.
Only three cases yielded an invalid result that was likely due to insufficient DNA (<0.5 ng/µl). Although this system has obtained the CE-IVD mark for FFPE material, the extracted DNA from routine smears can be directly pipetted into the cartridge, which may be automatically run as if an FFPE sample had been inserted. This off-label use of the Idylla test should not be seen as limiting factor since it is the same use of direct smears that classifies the procedure as a laboratory developed test, requiring in-house validation and quality control monitoring.
The time required to genotype DNA for KRAS mutational status assessment is approximately 2 h, compensating for the high cost related to the CE-IVD mark of a single cartridge (~€150). Although the process of removing the coverslip of archival smears does not compromise the quality of the DNA, it is time-consuming. In this study, smears archived for at least 2 years were employed, requiring even 3 days, but to avoid any delay, rapid on-site evaluation, at the time of the FNA procedure, enables the best triage of the sample for diagnosis and ancillary studies and the selection of a representative slide, that it is maintained uncoverslipped for immediate DNA extraction and Idylla processing.[1] Our results, showing that mCRC smears can reliably be tested by Idylla for KRAS mutation, and the recent availability of Idylla NRAS/BRAF test can give the cytopathologist the opportunity to sample metastatic sites to perform molecular analysis on cytological smears when histological resections are not available or when the patient is monitored to early detect the arising of mutant-resistant clones.[6]
CONCLUSION
This study demonstrates that the fully automated molecular diagnostics system Idylla™ is a promising opportunity for KRAS mutation testing on cytological samples. Even in less experienced laboratories, the cytopathologist may easily integrate morphological diagnostic report with accurate molecular information relevant for diagnostic and treatment decisions. However, it should be born in mind that long-term experiences are required to assess the feasibility of this automated molecular diagnostics system, and specific educational programs are required to enable pathologists to review and sign out clinical molecular genetic/molecular pathology results, and meanwhile, the relevance of a staff board-certified molecular pathologist or clinical molecular geneticist cannot be overemphasized.
COMPETING INTERESTS STATEMENT BY ALL AUTHORS
The authors declare that there is no relevant conflict of interest.
AUTHORSHIP STATEMENT BY ALL AUTHORS
All the authors read and approved the paper
ETHICS STATEMENT BY ALL AUTHORS
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with Helsinki Declaration of 1975, as revised in 2008.
LIST OF ABBREVIATIONS (In alphabetic order)
BRAF - v-raf murine sarcoma viral oncogene homolog B;
CE - IVD - In Vitro Diagnostic Medical Devices in Europe;
DNA - Deoxyribonucleic acid;
EGFR - Epidermal Growth Factor Receptor;
FFPE - Formalin Fixed Paraffin Embedded;
KRAS - Kirsten ras oncogene homolog;
mCRC - metastatic Colo – Rectal Cancer;
NRAS - Neuroblastoma RAS Viral Oncogene Homolog;
RT-PCR - Real Time Polimerase Chain Reaction.
EDITORIAL/PEER-REVIEW STATEMENT
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through automatic online system.
REFERENCES
- Applications and limitations of oncogene mutation testing in clinical cytopathology. Semin Diagn Pathol. 2013;30:284-97.
- [Google Scholar]
- Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with a KRAS mutation assay using allelic discrimination improves the diagnosis of pancreatic cancer. J Clin Gastroenterol. 2015;49:50-6.
- [Google Scholar]
- Cyst fluid: Moving beyond the carcinoembryonic antigen. Gastrointest Endosc. 2015;82:1070-1.
- [Google Scholar]
- KRAS mutation analysis on cytological specimens of metastatic colo-rectal cancer. Diagn Cytopathol. 2010;38:869-73.
- [Google Scholar]
- KRAS and BRAF mutation analysis can be reliably performed on aspirated cytological specimens of metastatic colorectal carcinoma. Cytopathology. 2011;22:358-64.
- [Google Scholar]
- Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532-6.
- [Google Scholar]
- Acquired KRAS mutations during progression of colorectal cancer metastases: Possible implications for therapy and prognosis. Cancer Chemother Pharmacol. 2010;66:605-9.
- [Google Scholar]
- Outsourcing cytological samples to a referral laboratory for EGFR testing in non-small cell lung cancer: Does theory meet practice? Cytopathology. 2015;26:312-7.
- [Google Scholar]
- Multi-center evaluation of the novel fully-automated PCR-based Idylla™ BRAF mutation test on formalin-fixed paraffin-embedded tissue of malignant melanoma. Exp Mol Pathol. 2015;99:485-91.
- [Google Scholar]
- BRAF mutation testing with a rapid, fully integrated molecular diagnostics system. Oncotarget. 2015;6:26886-94.
- [Google Scholar]
- Automated PCR detection of BRAF mutations in colorectal adenocarcinoma: A diagnostic test accuracy study. J Clin Pathol. 2016;69:398-402.
- [Google Scholar]
- Fully automated PCR detection of KRAS mutations on pancreatic endoscopic ultrasound fine-needle aspirates. J Clin Pathol 2016 pii: Jclinpath-2016-203696
- [Google Scholar]
- EGFR and KRAS mutations detection on lung cancer liquid-based cytology: A pilot study. J Clin Pathol. 2012;65:87-91.
- [Google Scholar]
- Role of gene expression profiling in defining indeterminate thyroid nodules in addition to BRAF analysis. Cancer Cytopathol. 2016;124:340-9.
- [Google Scholar]
- EGFR mutation detection on lung cancer cytological specimens by the novel fully automated PCR-based Idylla EGFR mutation assay. J Clin Pathol 2016 pii: Jclinpath-2016-203989
- [Google Scholar]
- Next generation sequencing improves the accuracy of KRAS mutation analysis in endoscopic ultrasound fine needle aspiration pancreatic lesions. PLoS One. 2014;9:e87651.
- [Google Scholar]
- Challenges and opportunities of next-generation sequencing: A cytopathologist's perspective. Cytopathology. 2015;26:271-83.
- [Google Scholar]








