Translate this page into:
Metallothionein 2A enhances the yes-associated protein 1 signaling pathway to promote small-cell lung cancer metastasis

*Corresponding author: Lidong Xu, Department of Cardiothoracic Surgery, The Second Hospital of Jiaxing, Jiaxing, China. 13456334819@163.com
-
Received: ,
Accepted: ,
How to cite this article: Wu H, Gu Y, Xu L. Metallothionein 2A enhances the yes-associated protein 1 signaling pathway to promote small-cell lung cancer metastasis. CytoJournal. 2025;22:25. doi: 10.25259/Cytojournal_201_2024
Abstract
Objective
Small-cell lung cancer (SCLC) remains challenging to treat due to its high invasiveness and propensity for drug resistance. Evidence suggests that the regulatory relationship between metallothionein 2A (MT2A) and the yes-associated protein 1 (YAP1) signaling pathway may influence the development of SCLC. Therefore, this study aims to explore the potential mechanisms affecting SCLC progression based on the regulatory interaction between YAP1 and MT2A.
Material and Methods
This study utilized reverse transcription quantitative polymerase chain reaction and Western blot analysis to analyze MT2A expression in cells. SCLC cell models with MT2A silencing and overexpression, as well as cotransfected cell models with YAP1 silencing and MT2A overexpression, were constructed. The effect of MT2A/YAP1 on cell growth, migration, and invasion was evaluated through a series of experiments, including cell viability assessment using cell counting kit-8 assay, colony formation examination, 5-ethynyl-2'-deoxyuridine staining, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling staining, and Transwell analysis. In addition, Western blot analysis was conducted to investigate alterations in crucial proteins associated with the YAP1 pathway and the epithelial-mesenchymal transition ( EMT) markers influenced by MT2A/YAP1. Lung metastasis and Ki67 expression were analyzed through hematoxylin and eosin staining and immunofluorescence analysis in vivo.
Results
In the SCLC cell line ( NCI-H69 cells), MT2A exhibits increased expression, facilitating cell growth, migration, and invasion. YAP1 expression decreases when MT2A is depleted. In addition, our findings validate that MT2A facilitates EMT progression and SCLC invasion and metastasis by upregulating YAP1 expression. In vitro, silencing MT2A inhibits lung metastasis and Ki67 expression.
Conclusion
MT2A facilitates the migration and invasion of SCLC cells by influencing the YAP1 signaling cascade. This investigation offers a fresh avenue for delving deeply into the potential mechanisms involved in the progression of SCLC.
Keywords
Metallothionein 2A
Small-cell lung carcinoma
Yes-associated protein 1
INTRODUCTION
Small-cell lung cancer (SCLC), known for its high aggressiveness and lethality among lung cancer subtypes, poses considerable treatment challenges.[1] Patients with SCLC have dismal therapeutic outcomes despite the substantial advancements in lung cancer treatment over the previous few decades, with their 5-year survival rates ranging from 5% to 10%.[2] This poor survival rate is largely attributed to SCLC’s highly invasive nature and propensity for early distant metastasis, making it difficult for conventional treatments to control disease progression effectively.
During cancer progression, the invasion and metastasis of tumor cells are among the primary reasons for treatment failure and recurrence.[3] Therefore, creating potent treatment plans requires a deepened understanding of the molecular mechanisms underlying SCLC, especially the signaling pathways controlling invasion and metastasis.
Metallothionein 2A (MT2A) is a crucial metal-binding protein that plays key roles in maintaining cellular homeostasis, redox reactions, and cell growth.[4,5] Recent studies have revealed aberrant expression levels of MT2A in various cancers, including lung cancer.[6-8] However, the specific role and mechanism of MT2A in SCLC remain unclear.
Another extensively studied signaling pathway is the Hippo pathway, which holds remarkable sway over cellular proliferation, apoptosis, and organ dimensions.[9] The aberrant activation of the Hippo signaling pathway has been closely linked to tumorigenesis and progression in numerous cancers, including lung cancer.[10,11] The Hippo signaling pathway’s major effector, yes-associated protein 1 (YAP1), is essential for controlling organ growth, apoptosis, and cell proliferation.[12] The dysregulation of the hippo pathway leads to increased YAP/transcriptional coactivator with PDZ-binding motif (TAZ) activity, which upregulates notch signaling components, promoting tumor growth and metastasis.[13]
Therefore, our study aims to elucidate the function of MT2A in SCLC and investigate its potential interactions with the YAP1 signaling pathway. Our goal is to delve deeply into the mechanisms underlying the contribution of this signaling pathway to the invasion and metastasis of SCLC, laying the groundwork for the development of innovative treatment strategies and therapeutic targets. Through this research endeavor, we aspire to offer renewed hope for the treatment and prognosis of patients with SCLC.
MATERIAL AND METHODS
Cell cultures
The SCLC cell line NCI-H69 (iCell-h472) and human bronchial epithelial cell line BEAS-2B (iCell-h023) were obtained from Cellverse Bioscience Technology Co., Ltd. (Shanghai, China). All cells were mycoplasma-free, and short tandem repeat analysis revealed that they were derived from their parental cells. The cells were cultivated separately in their corresponding culture media, which contained 10% fetal bovine serum (FBS, iCell-0500, iCell, Shanghai, China) and 1% P/S dual antibiotic (iCell-15140-122, iCell, Shanghai, China). Thaw the cryovial rapidly in a 37°C water bath, then transfer the cells to culture dishes with the prepared medium. As the cancer cells reached a certain density, cell passaging was performed to maintain their health. The cell suspension or adherent cells were placed in a constant-temperature and humidity cell culture incubator (MCO-5M, Panasonic, Osaka, Japan) and cultured at 37°C with 5% carbon dioxide. The culture medium was changed every 2–3 days to provide fresh nutrients and maintain a stable culture environment. The cells’ morphology, growth status, and purity were regularly monitored to ensure their health and purity.
Cell transfection
Negative control to MT2A (or YAP1) small interfering RNA (Si-NC, sense: 5'-UUCUCCGAACGUGUCACGUTT-3’, antisense: 5'-ACGUGACACGUUCGGAGAATT-3'), siMT2A (sense: 5'-GAGGAGAAGUAGAGCUGAUTT-3', antisense: 5'-AUCAGCUCUACUUCUCCUCTT-3'), siRNAYAP1 (sense: 5'-GGAGAUGAAUGCUGAAAGATT-3', antisense: 5'-UCUUUCAGCAUUCAUCUCCTT-3'), negative control to pCMV-MT2A, and pCMV-MT2A (sense: 5'-ATGGCGACCCGCACCCGCA-3', antisense: 5'- AAGCTTATGGCGCGCAGAGCAAGCAAACAG -3') were purchased from GenePharma (Shanghai, China). Plasmid vectors and siRNA oligonucleotides were transfected into NCI-H69 cells using Lipofectamine 3000 (L3000150, Invitrogen, Wilmington, Massachusetts, the USA). After 48 h of transfection, the cells were collected for transfection efficiency analysis through quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western blot analysis.
qRT-PCR
A commercial ribonucleic acid (RNA) extraction kit (R1100, Solarbio, Beijing, China) was used to extract RNA from the cells. The extracted RNA was transcribed into complementary deoxyribonucleic Acid (cDNA) using reverse transcriptase (18090200, Invitrogen, Wilmington, Massachusetts, the USA) and primers. The real-time polymerase chain reaction (PCR) reaction mixture, including PCR Master Mix (F631, MBI, Tokyo, Japan), primers (including specific primers and probes), and the transcription product (cDNA), was prepared and loaded into a real-time PCR instrument (CFX96, Bio-Rad, Hercules, California, the USA), and the PCR program was set up. The program included a series of temperature cycles, including denaturation, annealing, and extension steps. During real-time PCR, fluorescence signals were monitored and recorded for quantifying the amount of PCR product. The real-time PCR data were analyzed. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression was used as the endogenous control. Quantitative analysis was conducted using the 2−ΔΔCt method. The primer sequences used in this work are listed in Table 1.
| Primer name | Primer sequence (5'–3') |
|---|---|
| MT2A-F | AAGAGTGCAAATGCACCTC |
| MT2A-R | TCTTTACATCTGGGAGCGG |
| YAP1-F | TAGCCCTGCGTAGCCAGTTA |
| YAP1-R | TCATGCTTAGTCCACTGTCTGT |
| GAPDH-F | GGAGCGAGATCCCTCCAAAAT |
| GAPDH-R | GGCTGTTGTCATACTTCTCATGG |
MT2A: Metallothionein 2A, YAP1: Yes-associated protein 1, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Western blot analysis
Samples were added to centrifuge tubes containing protein extraction buffer, and ultrasonication was used to disrupt the cell membrane, hence releasing proteins. Proteins were extracted from the cells. The concentration of the extracted proteins was determined using the bicinchoninic acid assay (BCA) method (PC0020, Solarbio, Beijing, China). The extracted protein samples were loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS– PAGE) gel for electrophoresis separation in accordance with their molecular weight. The separated proteins from the SDS–PAGE gel were transferred onto a polyvinylidene fluoride membrane (YA1701, Solarbio, Beijing, China). Non-specific binding sites were blocked using a blocking buffer. Add the primary antibodies MT2A (1:1000 dilution, ab192385), E-cadherin (1:1000 dilution, ab314063), vimentin (1:1000 dilution, ab92547), Twist (1:1000 dilution, ab314056), large tumor suppressor 2 (LATS2, 1:1000 dilution, ab243657),phosph-YAP1 (1:1000 dilution, ab76252), YAP1 (1:1000 dilution, ab52771), TEA domain transcription factor 1(TEAD1, 1:1000 dilution, ab133535), and GAPDH (1:1000 dilution, ab9485, as the endogenous control) to the blocked membrane and incubated overnight at 4°C. The membrane was washed with tris-buffered saline with tween-20 (TBST) buffer to remove unbound primary antibodies. A horseradish peroxidase (HRP)-conjugated secondary antibody (1:1000 dilution, ab6721), which will bind to the primary antibody, was then added to the membrane. All antibodies were obtained from Abcam (Cambridge, MA, USA). The membrane was washed again with TBST buffer to remove the unbound secondary antibody. The immune complexes on the membrane were detected using enhanced chemiluminescence (BL520b, Biosharp Life Science, Hefei, Anhui, China) and a chemiluminescence imaging system (Image Quant LAS4000, GE Healthcare, Chicago, IL, the USA). The corresponding imaging system was then used for detection. Quantitative analysis was performed on gray-scale values using Image J software (version 1.48, National Institutes of Health, Rockville, MD, the USA). The gray-scale values were then normalized.
Cell counting kit-8 (CCK-8) assay
The cells to be tested were seeded into culture dishes and cultured under appropriate conditions to ensure stable growth. The culture dishes were then placed back into the cell culture incubator to continue cell culture under treatment conditions for a certain period. After 48 h of culture, CCK-8 reagent (CA1210, Solarbio, Beijing, China) was added to each treatment group. Following incubation, the cells were exposed to a culture medium supplemented with the CCK-8 reagent for 2 h. Subsequently, the optical density (OD) values of each treatment cohort were analyzed by utilizing a microplate reader (SpectraMax M2, Molecular Devices, San Jose, California, the USA) at 450 nm. Alterations in cell viability were evaluated by comparing variances.
Colony formation assay
The cells to be tested were cultured in culture dishes containing the appropriate culture medium and supplements to ensure their optimal growth condition. They were counted using a hemocytometer, and cell density was adjusted to 1 × 103 cells/dish. The culture dishes were placed in a constant-temperature incubator to allow the cells to adhere to the bottom of the dishes and form a monolayer. The cells were then further cultured in the incubator under stable conditions to allow growth and expansion. In accordance with the experimental design, the cells were cultured in the dishes for 7 days. The cells were stained with crystal violet for 10 min, and the stained cell colonies were visualized. The stained cell colonies were counted under a microscope (BX53, Olympus, Tokyo, Japan). Three fields of view were randomly selected. The differences in cell proliferation and growth under different experimental conditions were evaluated on the basis of the number and size of the colonies.
Transwell assay
Cell migration assay
Transwell membranes with appropriate pore sizes were inserted into a Transwell plate, and an appropriate amount of culture medium was added to each well. The cells to be tested were collected and resuspended in a culture medium. The treated cell suspension was added to the upper chamber of the Transwell plate. A certain number of cells per well were typically added. The even distribution of the cell suspension in the culture medium was ensured. To allow the cells to migrate from the upper chamber to the lower chamber, the Transwell plate was placed in a cell culture incubator, and the cells were cultured under the recommended conditions for approximately 2 h. After migration was complete, the Transwell membrane was removed, and non-migrating cells were gently wiped away from its upper surface with a cotton swab. The membrane was then stained with crystal violet (G1062, Solarbio, Beijing, China) staining solution for 20 min. Three fields of view were randomly selected. A microscope (BX53, Olympus, Tokyo, Japan) was used to observe the number of migrating cells.
Cell invasion assay
The Transwell membrane was coated with a layer of Matrigel to enhance the invasive ability of cells. Similar to the cell migration experiment, the cells to be tested were collected, resuspended in a culture medium, and treated as necessary. The treated cell suspension was added to the upper chamber of the Transwell, ensuring its even distribution in the culture medium. The Transwell plate was placed in a cell culture incubator, and the cells were cultured under appropriate conditions to allow invasive cells to migrate through the Matrigel layer and into the lower chamber. After the invasion was complete, the Transwell membrane was removed, and non-invasive cells were gently wiped away from its upper surface. The membrane was then stained with a dye. Three fields of view were randomly selected. A microscope (BX53, Olympus, Tokyo, Japan) was used to observe the number of invasive cells.
Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling staining
The cells were fixed at room temperature in 4% paraformaldehyde solution for 30 min, washed with phosphate buffer saline (PBS), soaked in Triton X-100 (P0096, Beyotime Biotechnology, Shanghai, China) for 5 min, and stained with 50 μL of TdT solution and 450 μL of fluorescein-labeled dUTP solution (D7428, Beyotime Biotechnology, Shanghai, China) for 60 min away from light. The cells were stained with 4',6-diamino-2-phenylindole (DAPI) for 15 min. Three fields of view were randomly selected. Images were observed with a fluorescence microscope (CX41-32RFL, Olympus Tokyo, Japan).
5-ethynyl-2'-deoxyuridine (EdU) staining
The cells were cultured in 96-well plates, and 1 × 104 cells were inoculated into each well. After overnight culture at 37°C, 100 μL of EdU (C0081, Beyotime Biotechnology, Shanghai, China) was added to each well. Incubation was continued at 37°C for 2 h. The cells were fixed with 4% paraformaldehyde at room temperature for 30 min and washed with PBS. Next, 100 μL of permeable solution (0.5% Triton X-100) was added to each well. Nuclei were stained with DAPI (C1005, Beyotime, Shanghai, China) and incubated at room temperature for 10 min away from light. Three fields of view were randomly selected. EdU-labeled cells were observed through fluorescence microscopy (CX41-32RFL, Olympus Corporation, Tokyo, Japan) and analyzed with ImageJ (v1.8.0.345, National Institutes of Health, Bethesda, MD, the USA).
Animal model of lung metastasis
Thirty male Balb/c nude mice weighing 18 ± 2 g and aged 4–6 weeks were used. All mice were provided by Beijing Vital River Laboratory Animal Technology Co., Ltd. (SCXK [Jing] 2016-0006). They were maintained under a 12 h light-dark cycle at 23°C ± 1°C and supplied free food and water. The mice were randomly divided into five groups, with six mice in each group, and injected with 200 μL of the cell suspension containing 5 × 106 SCLC through the tail vein.[14] The groups were designated as follows: Model (injection of SCLC), negative control to MT2A siRNA (Si-NC, injection of SCLC with negative control to MT2A siRNA), Si-MT2A (injection of SCLC with MT2A siRNA), negative control to pCMV-MT2A (Ov-NC, injection of SCLC pCMV-NC), and Ov-MT2A (injection of SCLC with pCMV-MT2A). After the experiment, the mice were killed through neck dislocation, and their lung tissues were collected. This study was approved by the ethics committee of our hospital. All animal procedures were performed in accordance with the guidelines for the care and use of animals.
Hematoxylin and eosin staining
The collected lung tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and sliced into 4 μm–thick sections. A hematoxylin and eosin staining kit (D006-1-1, Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China) was used, and the number of lung metastases was observed and analyzed using a microscope (BX46, Olympus, Tokyo, Japan).
Immunofluorescence staining
Lung tissue sections were treated with antigen repair solution and sealed at room temperature for 1 h with goat serum (I018-1-1 Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China). The tissues were washed with PBS twice and incubated with Ki67 (1:1000 dilution, ab15580, Abcam, Cambridge, MA, the USA) at 4°C overnight. The primary antibody was removed, and the tissues were washed with PBS and incubated with the secondary antibody (1:1000 dilution, ab150077, Abcam, Cambridge, MA, USA) at room temperature. After being washed with PBS, the tissues were incubated with DAPI (C0065, Solarbio, Beijing, China) for 15 min away from light. Finally, images were observed using a microscope (CX41-32RFL, Olympus Corporation, Tokyo, Japan), and fluorescence intensity was analyzed with ImageJ (v1.8.0.345, National Institutes of Health, Bethesda, MD, the USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 8.0.2, GraphPad Software, La Jolla, CA, USA). The results are presented as mean ± standard deviation and were derived from at least three independent experiments. Student’s t-test was employed to assess differences between two experimental groups, whereas analysis of variance with the Student–Newman–Keuls test was applied for comparisons involving three or more groups. Significance was defined as P < 0.05.
RESULTS
MT2A was upregulated in SCLC
By investigating the function of MT2A in SCLC, we scrutinized the expression patterns of MT2A in SCLC cells (NCI-H69 cells) and lung epithelial cells (BEAS-2B cells). The outcomes revealed that the messenger ribonucleic acid (mRNA) and protein levels of MT2A in NCI-H69 cells were significantly elevated compared with those in BEAS-2B cells (P < 0.01 and P < 0.001) [Figure 1a-c].
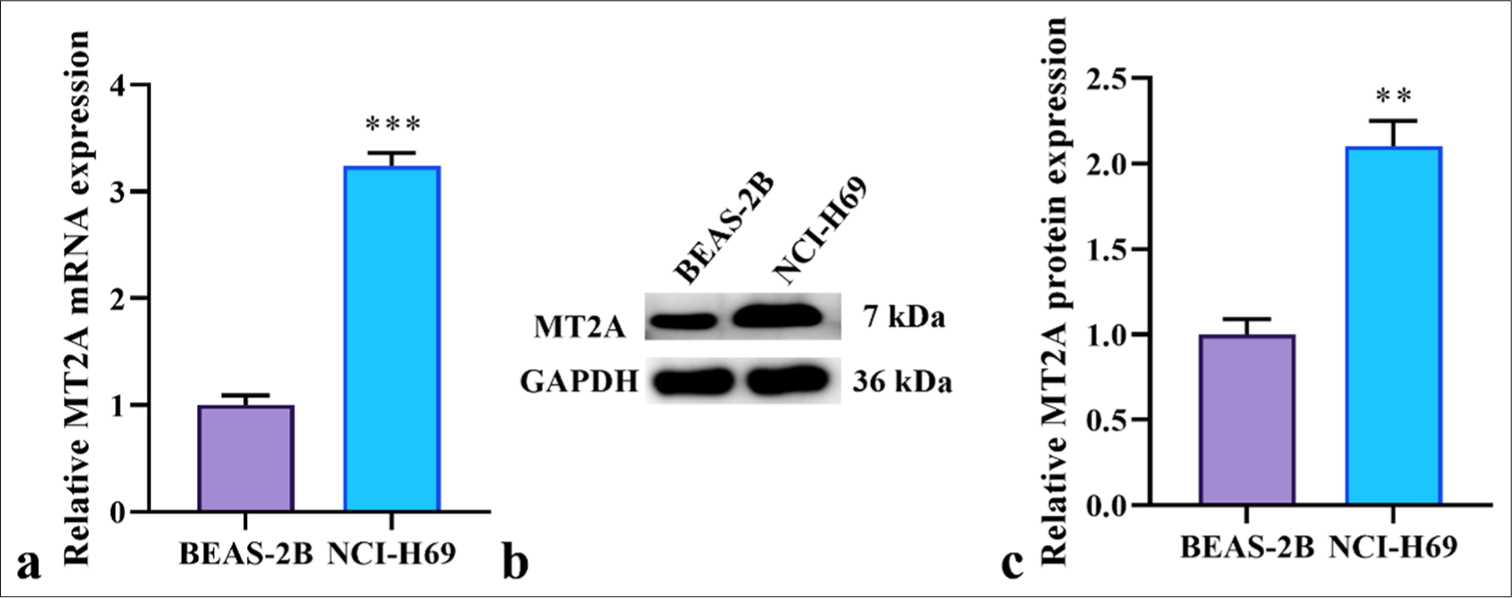
- Upregulation of MT2A in SCLC cells. (a) Expression of MT2A analyzed by RT-qPCR in SCLC cells (NCI-H69 cells) and lung epithelial cells (BEAS-2B cells). (b and c) Expression of MT2A analyzed by Western blot analysis in SCLC cells (NCI-H69 cells) and lung epithelial cells (BEAS-2B cells). n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. MT2A: Metallothionein 2A, SCLC: Small-cell lung cancer, GAPDH: Glyceraldehyde-3-phosphate dehydrogenase, RT-qPCR: Reverse transcription quantitative polymerase chain reaction.
MT2A promoted lung metastasis
We further demonstrated the role of MT2A in SCLC through animal experiments. Figures 2a and b show that MT2A silencing significantly reduced the number of lung metastases (P < 0.05), whereas MT2A overexpression significantly increased the number of lung metastases (P < 0.01). In addition, the immunofluorescence staining results of Ki67 in Figures 2c and d illustrate that the fluorescence intensity of Ki67 significantly reduced after MT2A was silenced but significantly increased after MT2A was overexpressed (P < 0.001).
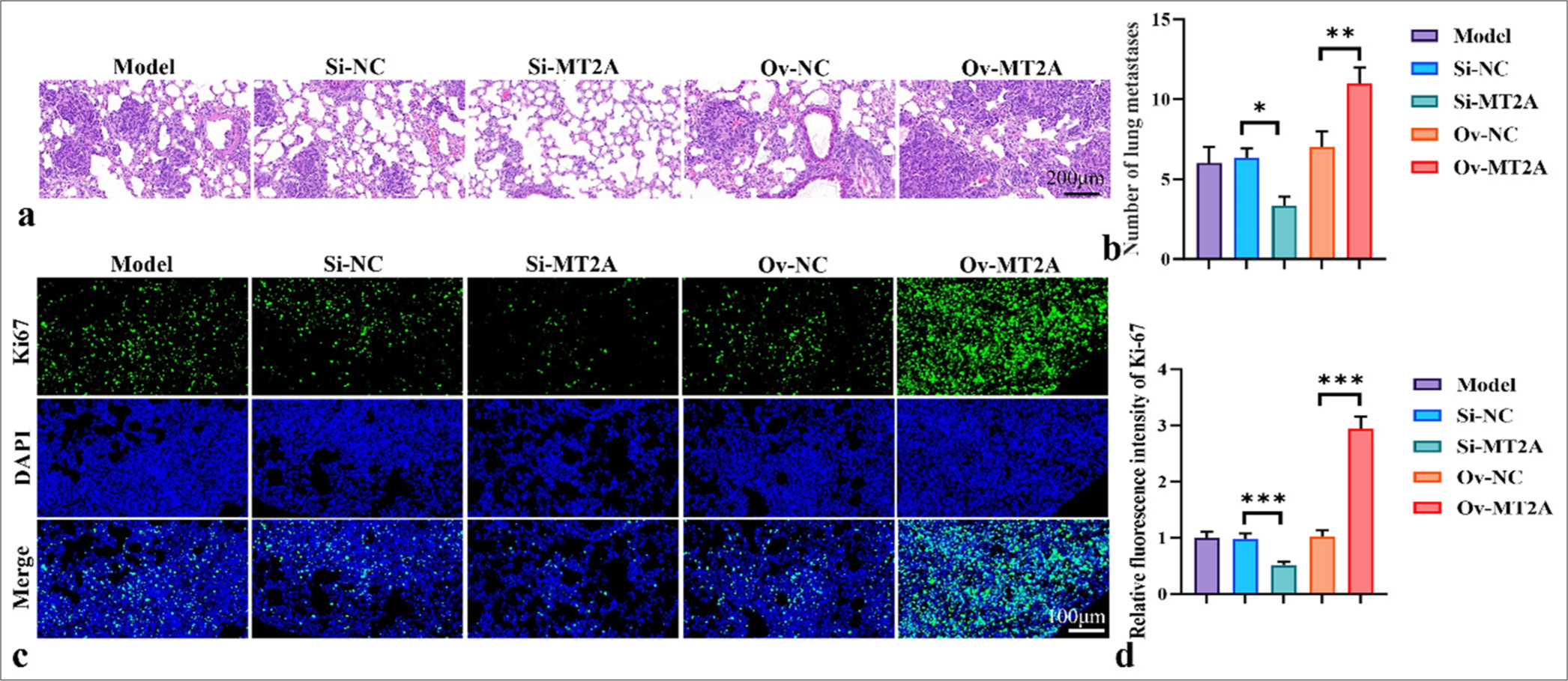
- MT2A promotes lung metastasis. (a and b) Representative image and histogram of lung metastasis. (c and d) Ki67 staining diagram and fluorescence intensity histogram, objective: 200×. n = 3, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. MT2A: Metallothionein 2A, DAPI: 4’,6-diamidino-2-phenylindole, Si-NC: negative control to MT2A (or YAP1) small interfering RNA, Ov-NC: negative control to pCMV-MT2A.
MT2A enhanced the migration and invasion of SCLC cells
We further explored NCI-H69 cells to investigate the involvement of MT2A in the cellular behavior of SCLC. Figure 3a-c illustrates the capacity of transfection with the MT2A siRNA and MT2A overexpression plasmids. The results revealed that MT2A expression was significantly downregulated following transfection with si-MT2A (P < 0.01). Conversely, MT2A expression was successfully upregulated after transfection with the MT2A overexpression plasmid (P < 0.001). Figure 3d-f demonstrates that MT2A downregulation notably impeded cell viability, whereas MT2A upregulation substantially enhanced cell proliferation (P < 0.001). Subsequently, we assessed the influence of MT2A on cell migration and invasion. Diminishing MT2A expression resulted in the inhibition of cell migration and penetration capability, whereas MT2A overexpression promoted cell migration and penetration (P < 0.001) [Figure 3g-j]. Finally, in NCI-H69 cells, MT2A overexpression significantly increased the proliferation rate and significantly decreased the apoptosis rate. In NCI-H69 cells, the proliferation rate significantly decreased, and the apoptosis rate significantly increased after MT2A silencing [Figure 3k-n] (P < 0.001).
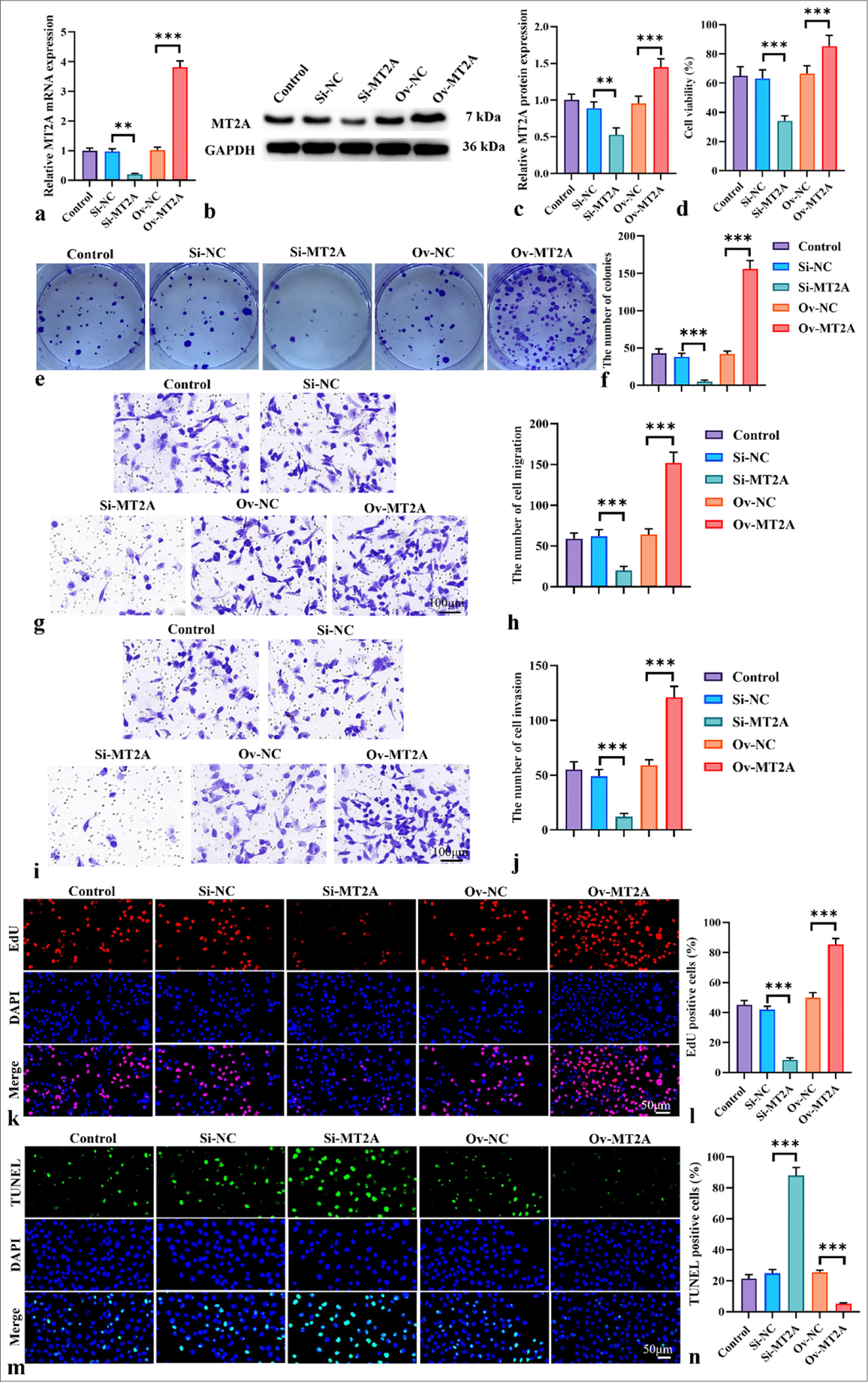
- MT2A enhances the invasion and migration of SCLC cells. (a-c) qRT-PCR and Western blot analysis, Objective: 200x, were used to analyze the efficiency of MT2A expression following the transfection of siRNA MT2A or pCMV-MT2A into NCI-H69 cells. (d) Proliferation of NCI-H69 cells after MT2A knockdown and overexpression was assessed by using the CCK-8 assay, Objective: 200x. (e and f) Proliferation of NCI-H69 cells after MT2A knockdown and overexpression was evaluated through the colony formation assay. (g-j) Transwell assay, Objective: 200x, was conducted to determine the migration (g and h) and invasion (i and j) of NCI-H69 cells after MT2A knockdown and overexpression. (k and l) Proliferation rate of NCI-H69 cells was determined through EdU staining, Objective: 200x. (m and n) Apoptosis rate of NCI-H69 cells was determined through TUNEL staining, Objective: 200x. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. MT2A: Metallothionein 2A, SCLC: Small-cell lung cancer, CCK-8: Cell counting kit-8, EdU: 5-ethynyl-2'-deoxyuridine, TUNEL: Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling, qRTPCR: Quantitative reverse transcription polymerase chain reaction.
MT2A enhanced EMT in SCLC cells
We examined the expression levels of EMT markers (E-cadherin, vimentin, and Twist1) in NCI-H69 cells following MT2A knockdown and overexpression to investigate the potential involvement of MT2A in the EMT of SCLC. Our findings showed that MT2A knockdown significantly increased E-cadherin protein levels while reducing the levels of vimentin and Twist1 protein production (P < 0.01) [Figure 4a-d]. By contrast, the overexpression of MT2A notably suppressed the protein expression levels of E-cadherin protein and enhanced those of vimentin and Twist1 (P < 0.001). These results indicate that the significant upregulation of MT2A promotes EMT in SCLC cells.
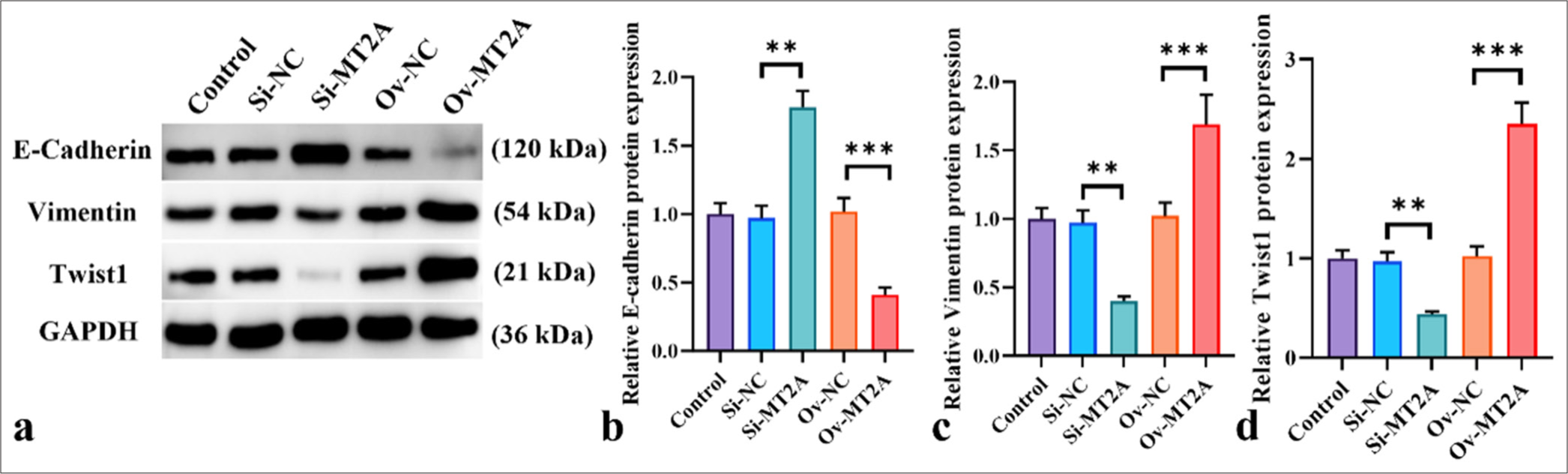
- MT2A facilitates EMT in SCLC cells. (a-d) Western blot analysis of E-cadherin, vimentin, and Twist1 following MT2A knockdown or overexpression. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. MT2A: Metallothionein 2A, SCLC: Small-cell lung cancer.
Knockdown of MT2A suppressed the activation of YAP1 signaling
The involvement of the YAP1 signaling pathway in SCLC development is notable. Western blot analysis indicated that in NCI-H69 cells, silencing MT2A with siRNA resulted in increased LAST2 protein expression levels, whereas MT2A overexpression led to a significant reduction in LAST2 protein expression levels (P < 0.05). In addition, the si-MT2A group demonstrated notably decreased protein expression levels of p-YAP1/YAP1 and TEAD1 (P < 0.01, and P < 0.001), whereas MT2A overexpression led to a significant increase in the expression levels of these proteins (P < 0.05, P < 0.01, and P < 0.001) [Figure 5a-d]. These findings suggest that MT2A may be able to inhibit the YAP1 signaling pathway. The results of the mRNA-level analysis provided in Figure 5e further demonstrate the link between MT2A and YAP1. Subsequently, we employed siRNA to silence YAP1 expression in NCI-H69 cells and concurrently cotransfected cells that were successfully transfected with siRNA-YAP1 with an MT2A overexpression plasmid. As depicted in Figure 5f-i, siRNA-YAP1 effectively suppressed YAP1 mRNA and protein expression in NCI-H69 cells, whereas OV-MT2A markedly elevated YAP1 expression (P < 0.01 and P < 0.001).
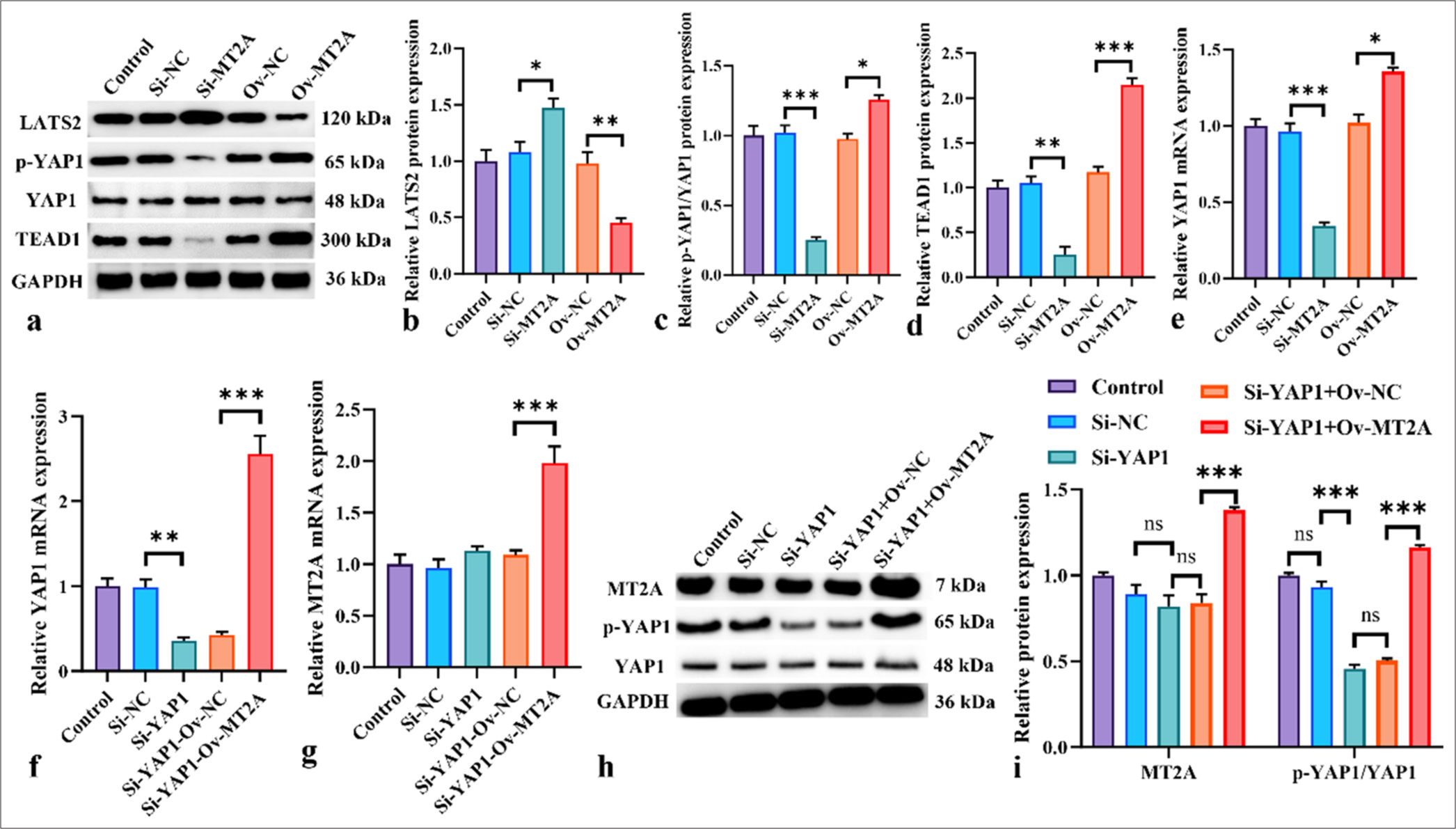
- Inhibition of YAP1 signaling activation by MT2A knockdown. (a-d) Western blot analysis was conducted to show the effects of MT2A overexpression or knockdown on TEAD, p-YAP1/YAP1, LAST2, and TEAD protein expression levels in NCI-H69 cells. (e) Effects of MT2A overexpression or knockdown on YAP1 mRNA levels. (f) YAP1 mRNA expression levels in NCI-H69 cells following YAP1 knockdown or cotransfection with the MT2A overexpression plasmid. (g) MT2A mRNA expression levels in NCI-H69 cells following YAP1 knockdown or cotransfection with the MT2A overexpression plasmid. (h and i) Expression levels of MT2A and p-YAP1/YAP1 after YAP1 silencing and MT2A overexpression. n = 3, ns: No significant difference, ✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001. YAP1: Yes-associated protein 1, MT2A: Metallothionein 2A, LATS2: Large tumor suppressor 2, TEAD1: TEA domain transcription factor 1.
MT2A promoted the proliferation, migration, and invasion of SCLC cells by regulating the expression of YAP1
Figure 6a-c demonstrates that YAP1 knockdown substantially diminished the viability of NCI-H69 cells, whereas MT2A overexpression counteracted the inhibitory effect of YAP1 knockdown on SCLC cell proliferation (P < 0.001). Transwell assays revealed that silencing YAP1 markedly hindered the migratory and invasive properties of NCI-H69 cells, effects that were mitigated by the overexpression of MT2A [Figure 6d-g]. In addition, in NCI-H69 cells, proliferation ability significantly decreased (P < 0.01), and apoptosis rate significantly increased (P < 0.01) after silencing YAP1. This trend was reversed after the overexpression of MT2A [Figure 6h-k]. In conclusion, the findings show that by stimulating the YAP1 signaling pathway, MT2A increases the survival and metastasis of SCLC cells. Finally, on the basis of the above conclusions, we speculate that the relationship between MT2A and YAP1 signaling pathways is shown in Figure 7.
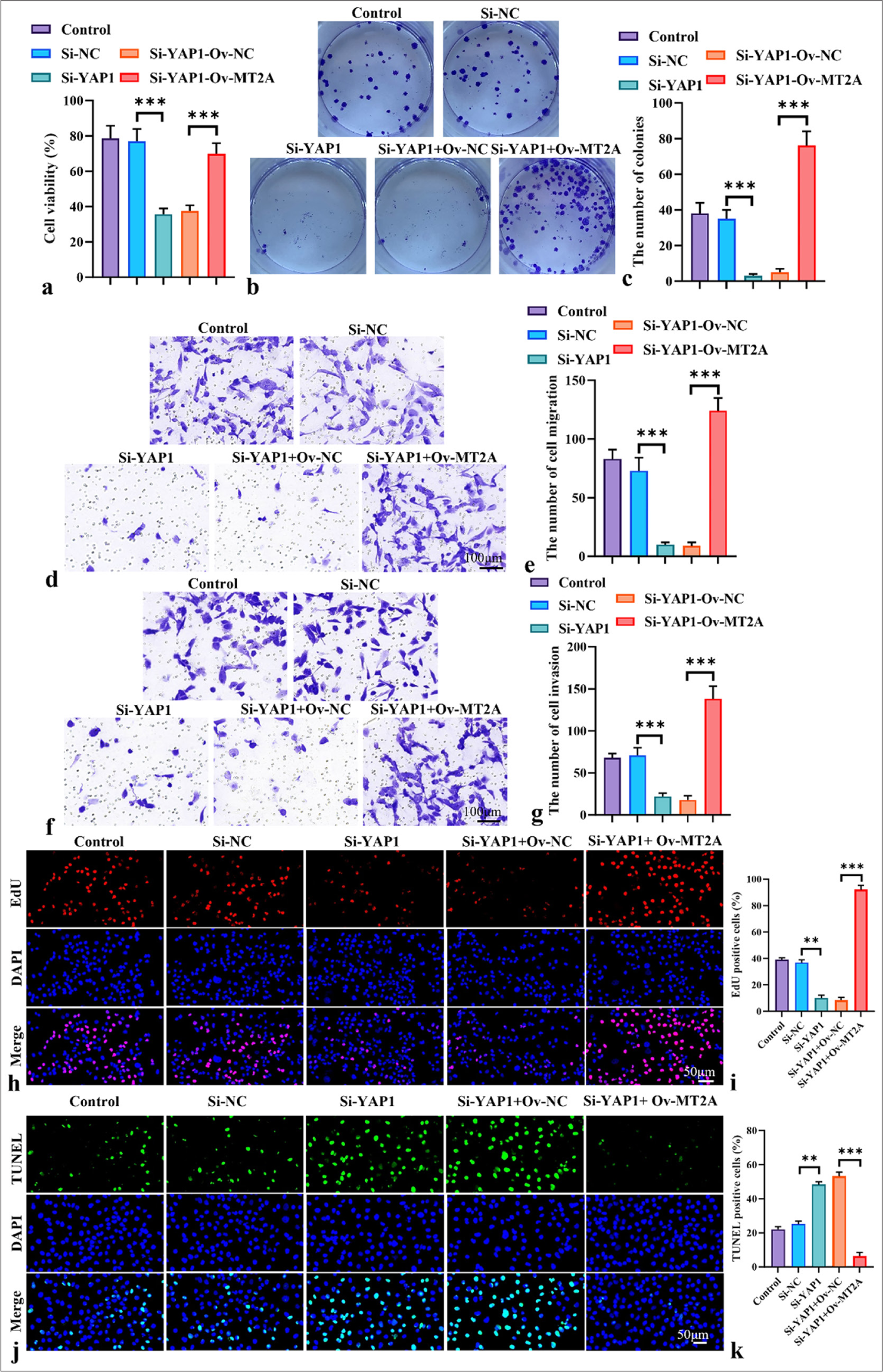
- MT2A promotes the proliferation and metastasis of SCLC cells by regulating YAP1 expression. (a) CCK-8 assay of the viability of NCI-H69 cells following YAP1 knockdown or cotransfection with the MT2A overexpression plasmid, Objective: 200x,. (b and c) Colony formation assay of the proliferation ability of NCI-H69 cells following YAP1 knockdown or cotransfection with the MT2A overexpression plasmid. (d-g) Transwell assay, Objective: 200x, analysis of the migration and invasion of NCI-H69 cells following YAP1 knockdown or cotransfection with the MT2A overexpression plasmid. (h and i) EdU staining, Objective: 200x, of NCI-H69 cells after YAP1 silencing and MT2A overexpression. (j and k) TUNEL staining, Objective: 200x, of NCI-H69 cells after YAP1 silencing and MT2A overexpression. n = 3, ✶✶P < 0.01, ✶✶✶P < 0.001. MT2A: Metallothionein 2A, SCLC: Small-cell lung cancer, YAP1: Yes-associated protein 1, CCK-8: Cell counting kit-8, EdU: 5-ethynyl-2'-deoxyuridine, TUNEL: Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling.
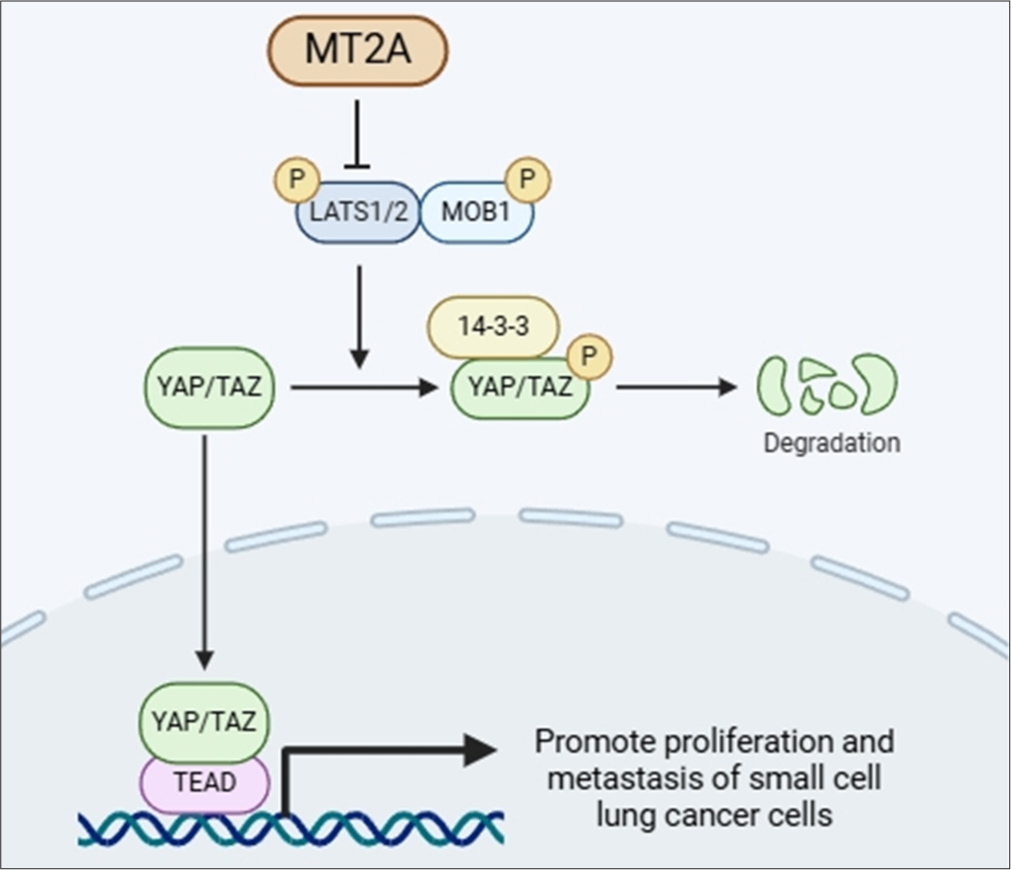
- Diagram of the connection between MT2A and the YAP1 signaling pathway drawn by using bioRender (https://www.biorender.com/). MT2A: Metallothionein 2A, YAP1: Yes-associated protein 1.
DISCUSSION
Our study reveals the important role of MT2A in SCLC and explores its potential interaction with the YAP1 signaling pathway. By observing the expression patterns and functional effects of MT2A, we uncover its potentially dual role in the onset and progression of SCLC.
Initially, we observed that MT2A was highly expressed in SCLC cells. In alignment with our research results, the findings of Wang et al. suggest that MT2A expression is upregulated in renal cell carcinoma.[15] MT2A is upregulated in gastric and breast cancers.[16,17] These results point to MT2A’s role as an oncogene. The proliferation, migration, and invasion capacities of SCLC cells were inhibited by MT2A knockdown, suggesting that MT2A may have a tumor-suppressive function in SCLC. However, we also found that MT2A overexpression promoted these cellular behaviors, suggesting the potential oncogenic role of MT2A. These findings are consistent with the results of some studies indicating that MT2A may have inhibitory effects in certain cancers but promoting effects in others.[18]
Further analysis revealed that MT2A may influence the migration and invasion capabilities of SCLC cells by regulating EMT. This result is consistent with previous research findings suggesting that MT2A may affect tumor invasion and metastasis by modulating cellular phenotypic transitions.[19]
In addition, we discovered a potential interaction between MT2A and the YAP1 signaling pathway that influences the biological behaviors of SCLC cells. This finding is novel and offers fresh insights into the mechanisms of action of MT2A in SCLC.
Regarding the molecular mechanisms between MT2A and the YAP1 signaling pathways, we speculate that MT2A may influence the activity of these pathways by directly or indirectly regulating the expression or activity of key proteins. For example, MT2A might interact with YAP1 and Tead1 in the Hippo pathway, affecting transcriptional regulation and signal transduction in cells.
Although our study sheds new light on the function of MT2A in SCLC, further investigation is required to confirm its results and examine any potential therapeutic uses. Future studies could elucidate the precise interactions between MT2A and the YAP1 signaling pathway through biochemical and cell biology experiments and investigate the clinical importance of MT2A as a potential therapeutic target and prognostic marker. In addition, we will repeat the experiment with multiple cell lines to further confirm the present results.
In summary, our study not only provides new insights into the role of MT2A in SCLC but also lays the foundation for a deepened understanding of its molecular mechanisms and potential clinical applications. Comparing the findings of our work with those of other studies is important.
SUMMARY
In SCLC, cell proliferation and metastasis are promoted by MT2A upregulation through the modulation of the YAP1 signaling pathway but are inhibited by MT2A downregulation.
AVAILABILITY OF DATA AND MATERIALS
The datasets and materials used and/or analyzed during the present study were available from the corresponding author upon reasonable request.
ABBREVIATIONS
CCK-8: Cell counting kit-8
DAPI: 4’,6-diamidino-2-phenylindole
ECL: Enhanced chemiluminescence;
EdU: 5-ethynyl-2’-deoxyuridine
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
H&E: Hematoxylin and eosin
MT2A: Metallothionein 2A
PVDF: Polyvinylidene fluoride
RT-qPCR: Reverse transcription quantitative polymerase chain reaction
SCLC: Small-cell lung cancer
SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresis
TUNEL: Terminal deoxynucleotidyl transferase-mediated
dUTP nick end labeling
YAP1: Yes-associated protein 1
ACKNOWLEDGMENT
Not applicable.
AUTHOR CONTRIBUTIONS
HW and YYG: Designed the study; all authors conducted the study; HW and LDX: Collected and analyzed the data; HW and LDX: Participated in drafting the manuscript, and all authors contributed to critical revision of the manuscript for important intellectual content. All authors gave final approval of the version to be published. All authors participated fully in the work, took public responsibility for appropriate portions of the content, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or completeness of any part of the work were appropriately investigated and resolved.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of The Second Hospital of Jiaxing (institution review board number, JXEY-ZFYJ108), dated 2024.8.30. All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of The Second Hospital of Jiaxing. Consent to participate was not required as this study did not involve human subjects.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING: Not applicable.
References
- Clinical insights into small cell lung cancer: Tumor heterogeneity, diagnosis, therapy, and future directions. CA Cancer J Clin. 2023;73:620-52.
- [CrossRef] [PubMed] [Google Scholar]
- Emerging Strategies for the treatment of small cell lung cancer: A review. JAMA Oncol. 2023;9:419-29.
- [CrossRef] [PubMed] [Google Scholar]
- The metastasizing mechanisms of lung cancer: Recent advances and therapeutic challenges. Biomed Pharmacother. 2021;138:111450.
- [CrossRef] [PubMed] [Google Scholar]
- Metallothionein 2A gene polymorphisms in relation to diseases and trace element levels in humans. Arh Hig Rada Toksikol. 2020;71:27-47.
- [CrossRef] [PubMed] [Google Scholar]
- Metallothionein 2A with antioxidant and antitumor activity is upregulated by caffeic acid phenethyl ester in human bladder carcinoma cells. Antioxidants (Basel). 2022;11:1509.
- [CrossRef] [PubMed] [Google Scholar]
- Metallothionein 2A expression in cancer-associated fibroblasts and cancer cells promotes esophageal squamous cell carcinoma progression. Cancers (Basel). 2021;13:4552.
- [CrossRef] [PubMed] [Google Scholar]
- Metallothionein 2A (MT2A) controls cell proliferation and liver metastasis by controlling the MST1/LATS2/YAP1 signaling pathway in colorectal cancer. Cancer Cell Int. 2022;22:205.
- [CrossRef] [PubMed] [Google Scholar]
- Esterase D interacts with metallothionein 2A and inhibits the migration of A549 lung cancer cells in vitro. J Cell Biochem. 2023;124:373-81.
- [CrossRef] [PubMed] [Google Scholar]
- The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin Sci (Lond). 2022;136:197-222.
- [CrossRef] [PubMed] [Google Scholar]
- CircXRN2 suppresses tumor progression driven by histone lactylation through activating the Hippo pathway in human bladder cancer. Mol Cancer. 2023;22:151.
- [CrossRef] [PubMed] [Google Scholar]
- The Hippo signaling pathway in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2023;55:893-903.
- [CrossRef] [PubMed] [Google Scholar]
- The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target ther. 2022;7:376.
- [CrossRef] [PubMed] [Google Scholar]
- Hippo pathway in regulating drug resistance of glioblastoma. Int J Mol Sci. 2021;22:13431.
- [CrossRef] [PubMed] [Google Scholar]
- The high expression of MTH1 and NUDT5 promotes tumor metastasis and indicates a poor prognosis in patients with non-small-cell lung cancer. Biochim Biophys Acta Mol Cell Res. 2021;1868:118895.
- [CrossRef] [PubMed] [Google Scholar]
- Single-cell transcriptome analysis revealing the intratumoral heterogeneity of ccRCC and validation of MT2A in pathogenesis. Funct Integr Genomics. 2023;23:300.
- [CrossRef] [PubMed] [Google Scholar]
- RING induces cell cycle arrest and apoptosis in human breast cancer cells by regulating the HSF1/MT2A axis. Exp Cell Res. 2023;433:113795.
- [CrossRef] [PubMed] [Google Scholar]
- ptk2 and mt2a genes expression in gastritis and gastric cancer patients with helicobacter pylori infection. Can J Gastroenterol Hepatol. 2022;2022:8699408.
- [CrossRef] [PubMed] [Google Scholar]
- Long-term 4-nonylphenol exposure drives cervical cell malignancy through MAPK-mediated ferroptosis inhibition. J Hazard Mater. 2024;471:134371.
- [CrossRef] [PubMed] [Google Scholar]
- PRKCSH alternative splicing involves in silica-induced expression of epithelialmesenchymal transition markers and cell proliferation. Dose Response. 2020;18:1-13.
- [CrossRef] [PubMed] [Google Scholar]








