Translate this page into:
Single-cell RNA sequencing analysis reveals the role of TXNDC5 in keloid formation


*Corresponding authors: Zhikun Liu, Department of Plastic Surgery, The Second Affiliated Hospital of Hainan Medical University, Haikou, Hainan, China. hnliuzhikun@126.com
Hongju Xie, Department of Plastic Surgery, The Second Affiliated Hospital of Hainan Medical University, Haikou, Hainan, China. Email: xhj1251@sina.com.
-
Received: ,
Accepted: ,
How to cite this article: Liu Z, Xian L, Li J, Zheng S, Xie H. Single-cell RNA sequencing analysis reveals the role of TXNDC5 in keloid formation. CytoJournal. 2024;21:40. doi: 10.25259/Cytojournal_58_2024
Abstract
Objective:
Thioredoxin domain-containing protein 5 (TXNDC5) is associated with fibrosis in a variety of organs, but its mechanism of action in keloid is unclear. In this study, we aimed to investigate the mechanism of TXNDC5 in keloid.
Material and Methods:
Single-cell RNA sequencing data of keloid and normal scar samples obtained from public databases were normalized and clustered using the Seurat package. Pathway enrich analysis was conducted using biological process enrichment analysis and Gene Set Enrichment Analysis (GSEA). In addition, TXNDC5 expression and its effects on migration and invasion of keloid fibroblasts (KFs) were validated based on cell function experiments.
Results:
A total of five cell types were obtained. The KF clusters were further clustered into two fibroblast subtypes (Fibroblast cells 1 and Fibroblast cells 2). Biological process enrichment analysis showed that transforming growth factor beta (TGF-β) signaling pathway was enriched in the two fibroblast subtypes. GSEA analysis demonstrated that genes in TGF-β signaling pathway were mainly enriched in Fibroblast cells 1, and that genes involved in cell proliferation, migration, and the TGF-β signaling pathway were all high-expressed in fibroblast cells 1. TXNDC5 was positively correlated with fibroblast proliferation, migration and TGF-β signaling pathway, and AUCell score. The cellular experiment confirmed that the messenger RNA and protein levels of TXNDC5 and TGF-β1 were high-expressed in KFs cells (P<0.001), and that knockdown of TXNDC5 downregulated TGF-β1 expression and inhibited migration and invasion of KFs (P<0.0001).
Conclusion:
Our study indicated that TGF-β signaling pathway was enriched in fibroblast cells, and TXNDC5 was positively correlated with proliferation, migration, and TGF-β signaling pathway. Cellular experiment demonstrated that knocking down TXNDC5 downregulated TGF-β1 expression, and suppressed migration and invasion of KFs. The current discoveries provided a new therapeutic strategy for the treatment of keloid.
Keywords
Keloid
Single-cell RNA sequencing
Fibroblast
Thioredoxin domain-containing protein 5
Transforming growth factor beta signaling pathway
INTRODUCTION
Keloid is a hyperproliferative skin response to dermal injuries such as trauma, surgery, burns, and inflammation.[1] Keloid persistently grows beyond the boundaries of the original wound and invades adjacent tissue structures, taking on the characteristics of a tumor as it can proliferate almost indefinitely.[2] The pathology of keloids is the excessive deposition of extracellular matrix [ECM] caused by abnormal proliferation of fibroblasts and secretion of matrix proteins.[3] Patients with keloids often experience pain and itching, which may seriously affect their psychological and physical quality of life.[4,5] Although some current clinical treatments, such as radiotherapy, surgery, interferon, and hormones, have improved the symptoms of keloid, the treatment efficacy of keloids is still limited due to a lack of comprehensive understanding on the pathogenic mechanisms of keloid.[6,7]
As a member of the protein disulfide isomerase (PDI) family, thioredoxin domain-containing protein 5 (TXNDC5) promotes the formation and rearrangement of disulfide bonds, which, in turn, supports proper protein folding.[8] A research have shown that TXNDC5 is aberrantly expressed in a variety of diseases, for example, upregulated TXNDC5 in cancer protects cancer cells from oxidative stress and promotes cancer cell growth, proliferation, migration, and angiogenesis.[9] In rheumatoid arthritis, again, TXNDC5 protects synovial fibroblasts from the deleterious effects of endoplasmic reticulum (ER) stress.[10] Furthermore, studies regarding lung fibrosis demonstrated that TXNDC5 can promote fiber formation through directly binding to transforming growth factor beta (TGF-β)1 in lung fibroblasts and stabilizing TGF-β1 signaling.[11] Similarly, a research have revealed the role of TXNDC5 in renal fibrosis through the TGF-β signaling pathway and identified the therapeutic potential of TXNDC5 targeting renal fibrosis .[12] Knockdown of TXNDC5 attenuates CCL4-induced hepatic fibrosis in mice through enhancement of ER stress.[13] TXNDC5 promotes tissue fibrosis by activating canonical (SMAD3-dependent) or non-canonical (MAP kinases [MAPK], such as c-Jun N-terminal kinase ( JNK), extracellular signal-regulated kinase (ERK) and survival protein STAT3) TGF-β signaling pathway.[14] It is known that the pathological nature of keloid is fibrosis of the dermal tissue.[15] However, the mechanism of action of TXNDC5 in keloid is still unknown.
A number of cell types have been identified as contributing to fibrosis, such as vascular endothelial cells, immune cells, and fibroblasts.[16] Fibroblasts are the central cell type in the process of skin fibrosis leading to extracellular matrix (ECM) accumulation and inflammation.[15,17] Fibroblasts in fibrotic diseases display excessive proliferative potential, increased migratory and invasive capacity, and increased ECM deposition that contribute to the pathogenesis of fibrosis.[15] These actions are primarily driven by fibrogenic growth factors such as TGF-β.[18,19] However, recent evidence suggests that fibroblasts are in fact a heterogeneous cell population in terms of morphology and function.[20,21] The development of single-cell RNA sequencing (scRNA-seq) provides an opportunity to study the heterogeneity of skin fibroblasts in homeostatic and pathological conditions. scRNA-seq suggested that fibroblasts in the normal human dermis are divided into several subgroups.[22,23]
Hence, in the present study, we hypothesized that TXNDC5 may promote keloid development and potentially promote keloid fibroblast (KF) activation and proliferation through TGF-β signaling pathway. The single-cell data of keloid from public databases were obtained to screen fibroblast subpopulations that could promote the cell proliferation, migration, and TGF-β signaling pathway. In addition, AUCell enrichment score was calculated to assess the molecular mechanisms involved in keloid development promoted by TXNDC5 through the TGF-β signaling pathway. The current discoveries contributed to the understanding of the pathogenesis and therapeutic targets of keloid, providing a new theoretical basis for the clinical treatment of patients with keloid.
MATERIAL AND METHODS
Acquisition of single-cell data
The scRNA-seq data of 3 keloid samples and 3 normal scar control samples were obtained from the gene expression omnibus (GEO) database (GSE163973, https://www.ncbi.nlm.nih.gov/geo/). Keloid sequencing libraries were constructed using ×10 Genomics and sequenced on Illumina NovaSeq 6000 system.
scRNA-seq data preprocessing and screening marker genes
The “Read10X” function in the Seurat package of R software was first carried out to read the scRNA-seq data of each keloid samples,[24] and then, single-cell data were normalized using “SCTransform” function. Subsequently, batch effects between keloid samples were removed by the harmony package. The uniform manifold approximation and projection (UMAP) was performed for downscaling based on the top 20 principal components and used the “FindNeighbors” and “FindClusters” functions to categorize the keloid cells. For clustering of all keloid cells, the resolution at 0.1 was set. Finally, the corresponding clusters of marker genes were annotated to specific cell types based on marker data in the CellMarker 2.0 database (http://bio-bigdata.hrbmu.edu.cn/CellMarker/).
Functional annotation of gene sets
The biological functions of a collection of genes of interest were explored by using the clusterProfiler package (version 4.8.2)[25] with the parameters of keyType = “ENTREZID,” pvalueCutoff = 0.05, and qvalueCutoff = 0.1.
A UCell enrichment analysis
The AUCell method could determine the activity status of gene collections in scRNA-seq data.[26] In this regard, gene data of the TGF-β signaling pathway were downloaded from the Kyoto Encyclopedia of Genes and Genomes (https://www. genome.jp/kegg/), and obtained gene data related to fibroblast proliferation, migration, apoptosis, and the TGF-β signaling pathway from the MsigDB database (https://www.gsea-msigdb. org/gsea/msigdb). Subsequently, expression matrices for fibroblasts were extracted and AUCell scores were calculated for these sets of genes within each sample using the AUCell package.
Cell culture and transfection
The KEL FIB (KF, cat. CRL-1762) cells and normal fibroblasts (NFs, cat. PCS-201-010) were purchased from American Type Culture Collection (ATCC) (Manassas, USA) and cultured in Dulbecco’s modified Eagle medium (DMEM, M4655, Merck, Darmstadt, Germany) containing 10% fetal bovine serum (S9020, Solarbio, China) in the cell incubator containing 5% CO2 at 37°C. Cells have been STR identified and the mycoplasma detection results are negative. The cells were treated with diluted small interfering (si) RNA targeting TXNDC5 (si-TXNDC5#1: 5’-ATCGAGCTACTTCCCATAATA-3’; si-TXNDC5#2: 5’-AGGGCCCTAACTAGAGTTCTA-3’.) and negative control (si-NC: 5'-UUCUUCGAACGU GUCACGUTT-3') purchased from GenePharma (Suzhou, China). The treatment was performed using Lipofectamine® 3000 (L3000150, Thermo Fisher Scientific, Carlsbad, CA, USA), following the manufacturer’s guidelines. Transfection was conducted using serum-free DMEM for 8 h, the serum-depleted DMEM was substituted with DMEM supplemented with 10% fetal bovine serum to continue the cell incubation for another 48 h.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
After the cell transfection, the cells were collected and total RNA (1 mg) was extracted with the GoScript Reverse Transcription System kit (A5003, Promega Corporation, Madison, Wisconsin, USA). After quantification and synthesis of cDNA, quantitative polymerase chain reaction was performed using SYBR Green Master Mix (1708880, Bio-Rad, USA). The primers used were as follows:
TXNDC5 5’- CAGAGCCGGAA
GTGGAACC-3’ (forward), 5’- CCACGGAGCGA
AGAACTTGAT-3’ (reverse); TGF-β1 5’- GGCCAGATC
CTGTCCAAGC-3’ (forward), 5’- GTGGGTTTCCA
CCATTAGCAC-3’ (reverse); Smad2, 5’-CCG AAATGCC
ACGGTAGAAA-3’ (forward) and 5’-GGGCTCTGCACAAAGATTGC-3’ (reverse); Smad3, 5’-CCCCAGCACATAAT
AACTTGG-3’ (forward) and 5’-AGGAGATGGAGCA
CCAGAAG-3’ (reverse). β-actin was a housekeeping gene, 5’-CCTTCCTGGGCA
TGGAGTCCT-3’(forward), and 5’-AATCTCATCTTG
TTTTCTGCG-3’ (reverse). Furthermore, the relative quantification of genes was calculated using the 2−ΔΔCt method[27].
Western blot
Proteins were obtained from normal fibroblasts (NFs) and KEL FIB (KFs) using radioimmunoprecipitation assay (RIPA) (R0010, Solarbio, Beijing, China). The protein concentration of cell lysates was determined using a bicinchoninic acid (BCA) protein assay kit (E112-01, Vazyme, Nanjing, China). 30 mg of protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electrotransferred to polyvinylidene difluoride membranes (PVDF, IPVH00010, MILLIPORE, Billerica, Massachusetts, USA). The membranes were blocked with 5% skimmed milk, washed with Tris-buffered saline with Tween-20 (TBST, T1081, Solarbio, Beijing, China) buffer, and then washed with primary antibodies TGF-β1 (ab315254, 1:1000, Abcam, Cambridge, United Kingdom) and TXNDC5 (ab13820, 1:1000, Abcam, Cambridge, United Kingdom), overnight at 4°C. The membranes treated with primary antibodies were, then, washed 3 times with TBST and incubated with anti-rabbit enzyme-linked antibody for 1 h at room temperature, with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference. Signals were measured by Immobilon Western Chemiluminescent HRP substrate (Millipore Corporation, Billerica, MA, USA) and an Amersham Imager 600 imager (GE healthcare Life science, Pittsburgh, USA) for visualization. Regarding Western Blot quantification, normalized was performed.
Transwell assay
The KF cells invasion was assessed using Transwell chambers with 8 mm pore size and pre-coated with Matrigel (Corning, Inc. USA). The transfected KF cells (1 × 104 cells/well) contained in serum-free DMEM were seeded into the upper chambers. Following 48-h incubation, the medium was removed, and invading cells were fixed in ice-cold 100% methanol, stained with 0.1% crystal violet (G1063, Solarbio, China) for 3 min, and washed with Phosphate-Buffered Saline (PBS P1000, Solarbio, Beijing, China). Then, the cells from three random fields were counted under a microscope (Eclipse Ts2, Nikon, Tokyo, Japan).
Wound-healing assay
The transfected KF cells were placed in a 6-well plate and the center of the cell monolayer was scratched with a pipette tip. Subsequently, the cells were washed 3 times with PBS and cultured in serum-free DMEM. Images of the wounds were taken under a light microscope (magnification, ×100) at 0 h and 48 h, respectively. Finally, the width of the wounds was measured using Image J software (National Institutes of Health, Bethesda, Maryland, USA) to assess the migratory capacity of the KFs. The wound healing rate was calculated as [(0 hr width - 48 hr width)/0 hr width] × 100%.
Statistical analysis
R software (version 4.3.1, Columbia University Irving Medical Center, New York, NY, USA) and GraphPad Prism software (version 8.0.2, GraphPad, LLC., La Jolla, California) were utilized for all computational and analytical analyses in this research investigation. Student’s t-test was used to compare the differences in continuous variables between the two sample groups. The experimental results were obtained from three separate experiments. Statistical significance was determined under a threshold of P < 0.05.
RESULTS
Single-cell landscape analysis of keloids
Seurat package was used to normalize, downscale, and cluster the scRNA-seq data of keloid with cellular annotation to obtain five cell clusters, including fibroblast cells, epidermal stem cells, endothelial cells, macrophages, and mast cells [Figures 1a and Figure S1]. Specifically, KF cells high-expressed THY1, TAGLN, MYH11, FGF7, ASPN, GPC3, SFRP1, WISP2, and MFAP5; endothelial cells high-expressed FLT1, KDR, VWF, PECAM1, and CDH5; epidermal stem cells high-expressed KRT5, KRT14, TP63, KRT1, SBSN, and KRTDAP; macrophages high-expressed CD163, CSF1R, CYBB, and FPR3; and mast cells high-expressed SLC18A2, CPA3, HPGDS, and TPSB2 [Figure 1b and c]. Finally, the proportion of different cell types in the keloid were determined. As shown in Figure 1d and 1e, the percentage of fibroblast cells was the highest in keloid patients (43.22%), which predicted a potential influence of fibroblasts cells in the development of keloid. However, since the pathological nature of keloid was the fibrosis of dermal tissue, the fibroblast clusters in keloid was explored.
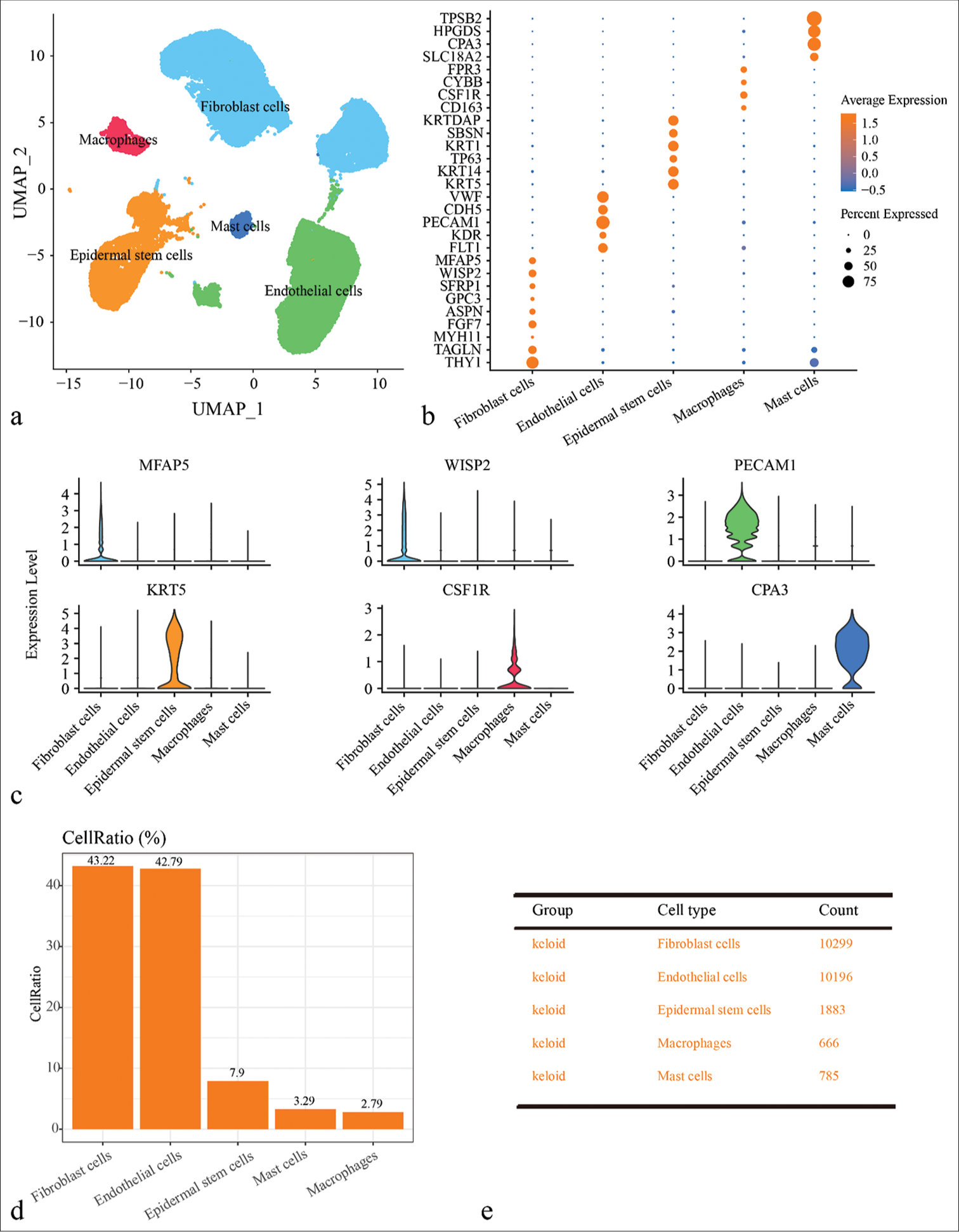
- Single-cell landscape of keloid and normal scar samples revealed by single-cell RNA sequencing (scRNA-seq) data. (a) Uniform manifold approximation and projection plot showing scRNA-seq data annotated for five different cell types, including fibroblast cells, epidermal stem cells, endothelial cells, macrophages, and mast cells. (b) Bubble plots of expression levels of marker genes in different cell types. (c) Violin diagram showing expression of major marker genes in different cell types. (d) Bar plot showing the proportion of cells of different cell types in different samples. (e) Demonstration of cell numbers of different cell types in keloid and normal scar groups. (UMAP: uniform manifold approximation and projection; TPSB2: tryptase beta 2; HPGDS: Hematopoietic prostaglandin D synthase; CPA3: carboxypeptidase A3; SLC18A2: solute carrier family 18 member A2; FPR3: formyl peptide receptor 3; CYBB: cytochrome b-245 beta chain; CSF1R: colony stimulating factor 1 receptor; KRTDAP: keratinocyte differentiation associated protein; SBSN: suprabasin; KRT1: keratin 1; TP63: tumor protein p63; KRT14: keratin 14; KRT5: keratin 5; VWF: von Willebrand factor; CDH5: cadherin 5; PECAM1: platelet and endothelial cell adhesion molecule 1; KDR: kinase insert domain receptor; FLT1: FMS-like tyrosine kinase 1; MFAP5: microfibril associated protein 5; WISP2: WNT1 inducible signaling pathway protein 2; SFRP1: secreted frizzled related protein 1; GPC3: glypican 3; ASPN: asporin; FGF7: fibroblast growth factor 7; MYH11: myosin heavy chain 11.)
Characterization of fibroblasts in keloid
To further understand the potential role of fibroblasts in Keloid, all the single-cell data of fibroblasts were extracted for clustering and identified two subpopulations of fibroblasts, including fibroblast cells 1 and fibroblast cells 2 [Figure 2a]. In addition, the biological functions of fibroblast subpopulations in keloid were analyzed and it is found that fibroblast cells 1 were mainly enriched in pathways such as the Wnt signaling, angiogenesis, and epithelial cell proliferation and migration, while fibroblast cells 2 was mainly enriched in pathways such as MAPK signaling pathway, Rho protein signal transduction, and wound healing. Both the fibroblast subpopulations were enriched in the relevant TGF-β signaling pathway [Figure 2b]. Next, gene expressions in the two fibroblast subpopulations were explored and it is observed that LUM, COL3A1, COL1A2, COL1A1, FBLN1, and SFRP2 were high-expressed in fibroblast cells 1, while PDGFA, MT1A, TINAGL1, MCAM, ID4, and RGS5 were present with higher expression in fibroblast cells 2 [Figure 2c]. Gene set enrichment analysis results indicated that genes in the TGF-β signaling pathway were enriched in the fibroblast cells 1 [Figure 2d]. In addition, [Figure S2] showed that genes involved in cell proliferation, migration, and the TGF-β signaling pathway were more high-expressed in fibroblast cells 1. In particular, the AUCell enrichment score of TGF-β signaling pathway were higher in Keloid and fibroblast cells 1, suggesting that fibroblasts played an important role in keloid, which was likely to be regulated by TGF-β signaling pathway [Figure 2e and f].
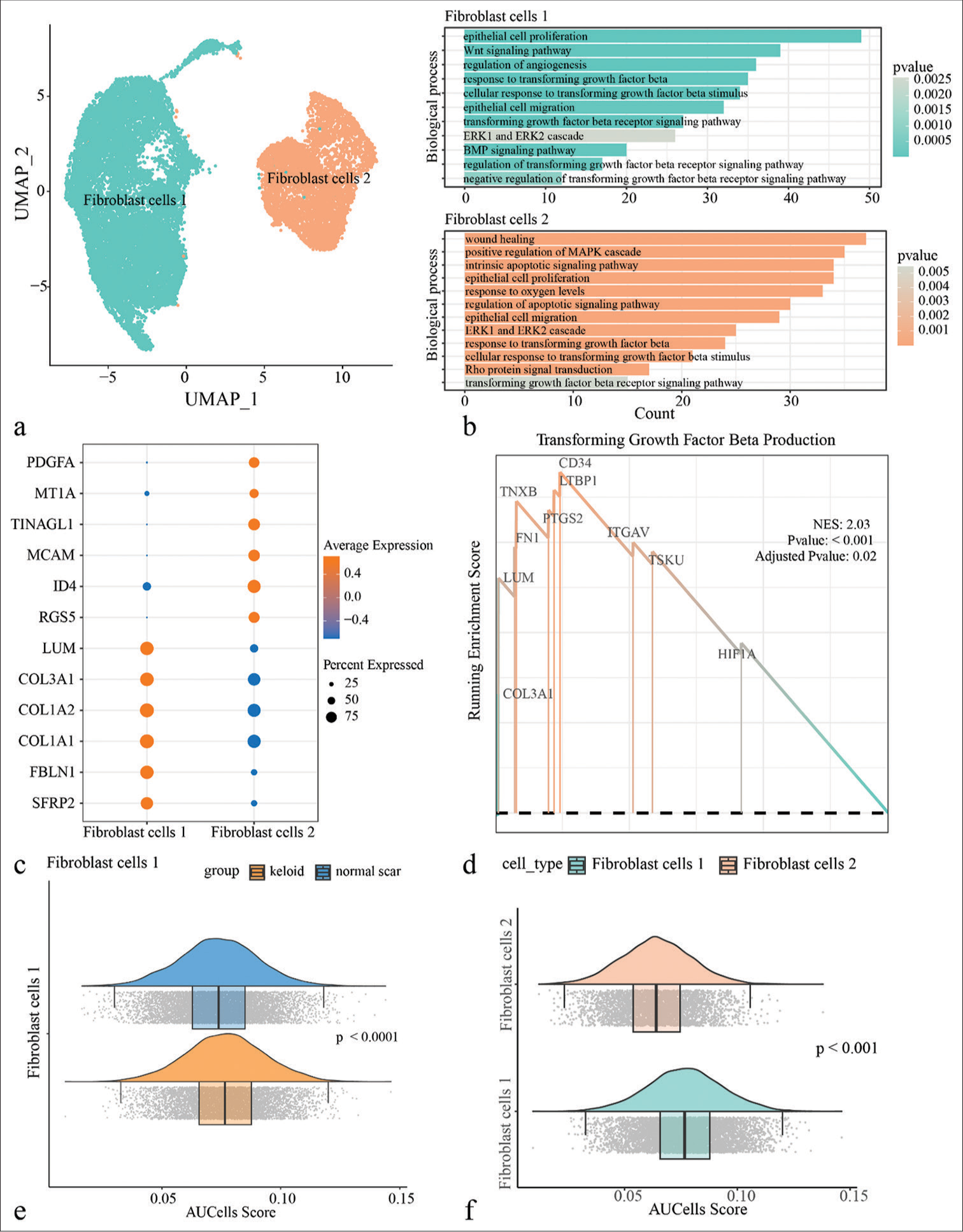
- Single-cell landscape of fibroblasts in the keloid. (a) All keloid fibroblasts were clustered by uniform manifold approximation and projection and divided into two subpopulations, fibroblast cells 1 and fibroblast cells 2. (b) Enrichment analysis of biological processes of Differentially expressed genes (DEGs) between different fibroblast subpopulations. (c) Demonstration of highly expressed genes in two fibroblast subpopulations. (d) Gene set enrichment analysis results demonstrate that genes in the transforming growth factor-beta (TGF-β) signaling pathway are predominantly concentrated in fibroblast cells 1. (e) AUCell enrichment analysis of fibroblast cells 1 between keloid and normal scar in the TGF-β signaling pathway. (f) AUCell enrichment analysis of fibroblast cells 1 and fibroblast cells 2 in the TGF-β signaling pathway. (UMAP: uniform manifold approximation and projection; PDGFA: platelet derived growth factor subunit A; MT1A: metallothionein 1A; TINAGL1: tubulointerstitial nephritis antigen-like 1; MCAM: melanoma cell adhesion molecule; ID4: inhibitor of DNA binding 4; RGS5: regulator of G-protein signaling 5; LUM: lumican; COL3A1: collagen type III alpha 1 chain; COL1A2: collagen type I alpha 2 chain; COL1A1: collagen type I alpha 1 chain; FBLN: fibulin 1; SFRP2: secreted frizzled related protein 2; TXNB: thioredoxin b; FN1: fibronectin 1; PTGS2: prostaglandin-endoperoxide synthase 2; ITGAV: integrin subunit alpha V; TSKU: tsukushi, small leucine rich proteoglycan; HIF1A: hypoxia inducible factor 1 subunit alpha.)
Knockdown TXNDC5 downregulated TGF-β1 expression and inhibited migration and invasion of KFs cells
Based on the previous studies, TXNDC5 was involved in the formation of fibrosis in multiple organs. Therefore, the correlation between expression of TXNDC5 and fibroblasts and TGF-β signaling pathway was first explored, and observed that the TXNDC5 gene was significantly positively correlated with AUCell score of cell proliferation, migration, and TGF-β signaling pathway in keloid [Figure 3]. From bulk-RNA-Seq analysis, GES158395, GSE188952, and GSE190626 dataset were combined into a whole dataset containing 11 keloid and 15 normal scars. Results showed that TXNDC5 had higher expression in keloid in comparison to normal scars [Figure S3a]. Moreover, a positive phenomenon was observed between TXNDC5 expression and proliferation, migration, apoptosis, and TGF-β signaling pathway [Figure S3b-e]. [Figure S4a and b] also presented different pathways enriched in high TXNDC5 expression group and low TXNDC5 expression group. Those data indicated that TXNDC5 maybe involve in the progression of keloid.
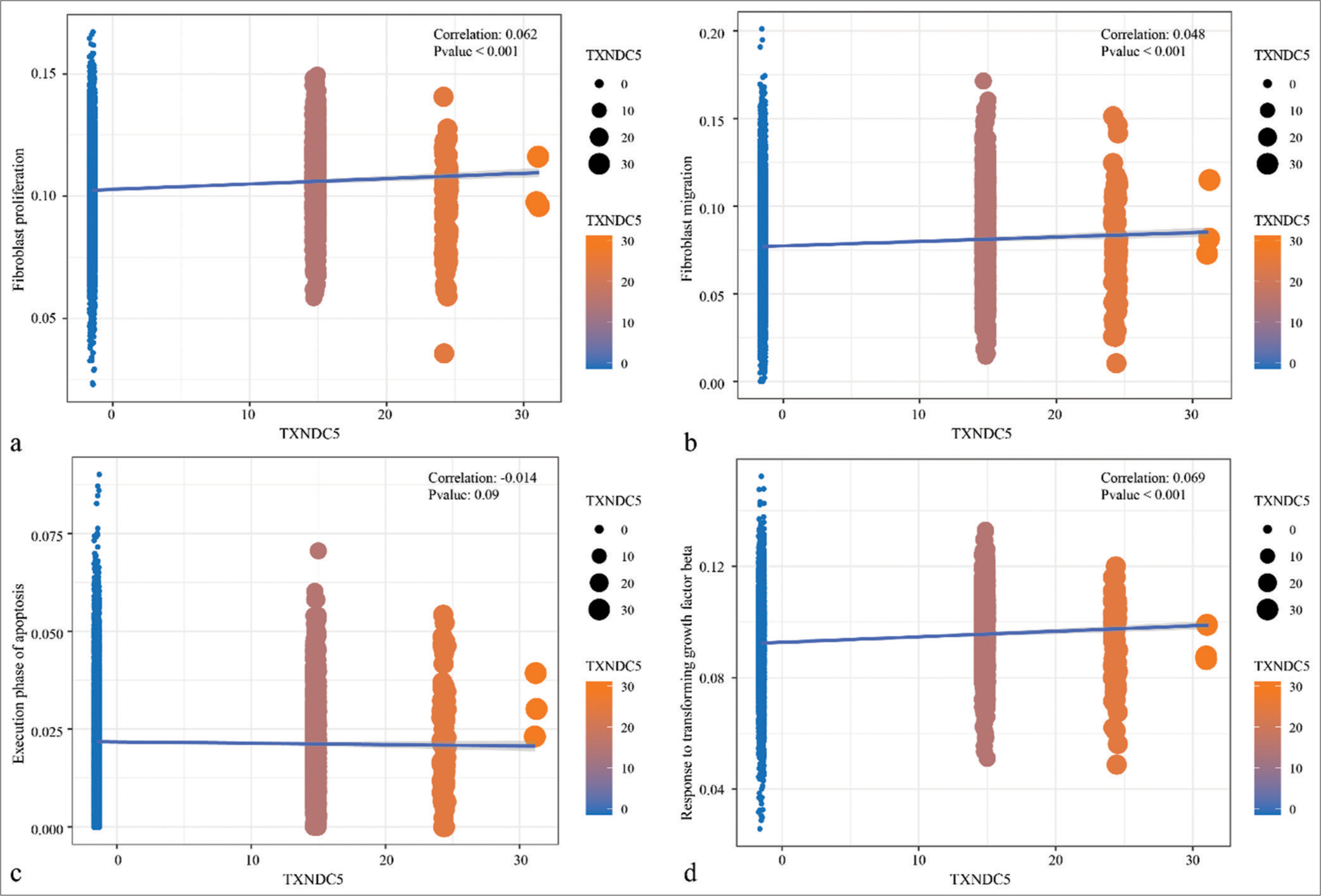
- Correlation analysis between thioredoxin domain-containing protein 5 gene expression levels and pathways. (a) Correlation analysis of thioredoxin domain-containing protein 5 gene expression levels with AUCell score of genes in the fibroblast proliferation. (b) Correlation analysis of thioredoxin domain-containing protein 5 gene expression levels with AUCell score of genes in the fibroblast migration. (c) Correlation analysis of thioredoxin domain-containing protein 5 gene expression levels with AUCell score of genes in the execution phase of apoptosis. (d) Correlation analysis of thioredoxin domain-containing protein 5 gene expression levels with AUCell score of genes in the response to transforming growth factor-beta signaling pathways.
Subsequently, the expression levels of TGF-β1 and TXNDC5 in KFs were validated based on RT-qPCR. The results showed that the expression levels of TGF-β1 and TXNDC5 were significantly higher in KFs compared to normal fibroblasts (NFs) [P<0.001, Figure 4a]. Western blot results also showed that protein expressions levels of TGF-β1 and TXNDC5 were upregulated in KFs cells [P<0.001, Figure 4b]. Results of RTqPCR showed that the messenger RNA (mRNA) expressions of Smad2 and Smad3 were upregulated in KFs compared to NFs [P<0.05, Figure 4c], which were corresponding with the increased activation of TGF-β signaling pathway in KFs. Next, siRNA TXNDC5 was used to downregulate TXNDC5 expression in KFs [P<0.001, Figure 4d] and found that the knockdown TXNDC5 lowered the protein expression and mRNA level of TGF-β1 [P<0.05, Figure 4e and f]. The effect of TXNDC5 on KF migration and invasion was evaluated using the Transwell invasion and wound healing assay. The results demonstrated that silencing TXNDC5 expression reduced the invasive ability of KFs [P<0.0001, Figure 4g]. Similarly, the results of the wound healing assay showed that inhibition of TXNDC5 expression suppressed the migratory ability of KF cells [P<0.0001, Figure 4h].
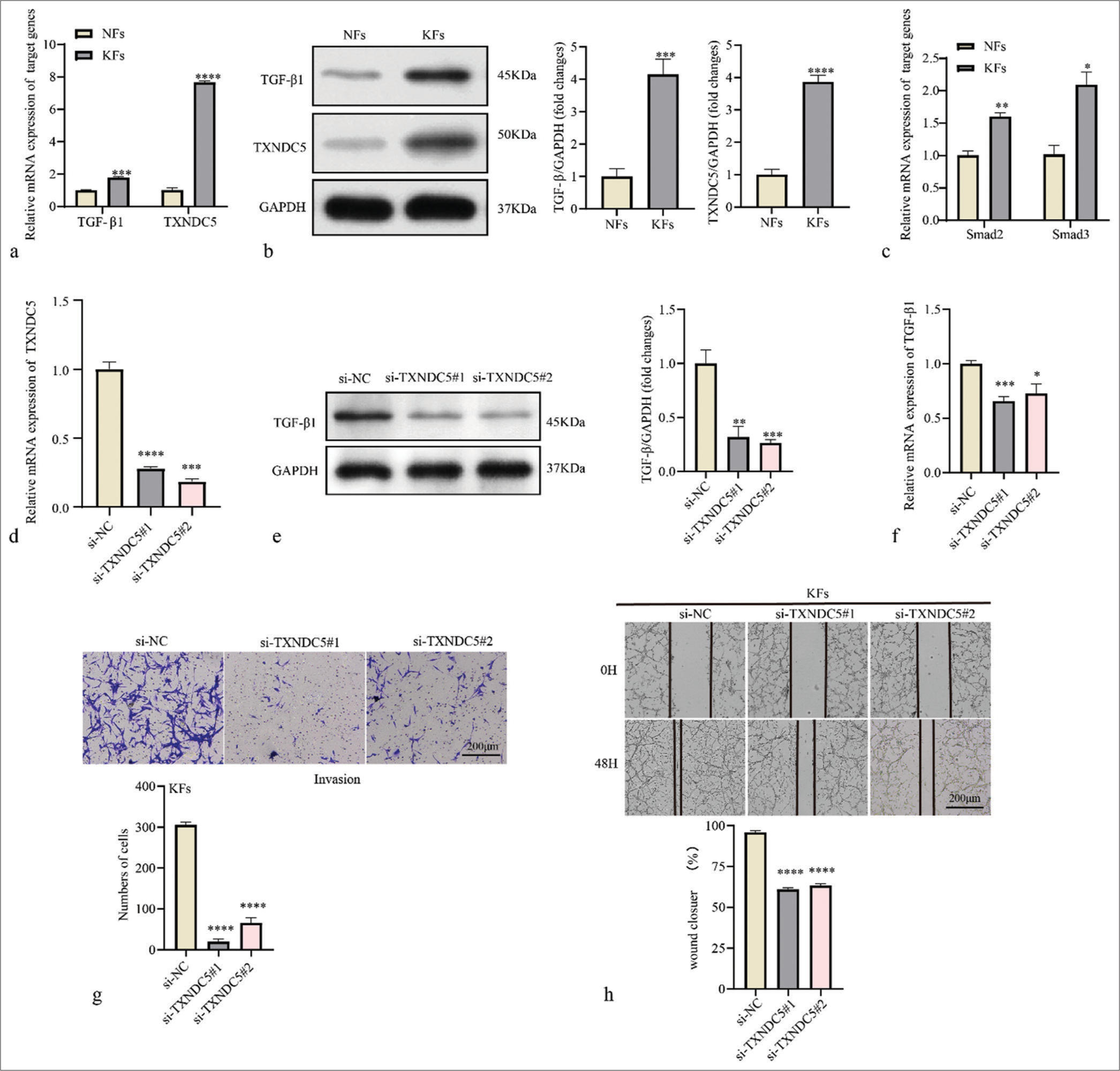
- Thioredoxin domain-containing protein 5 (TXNDC5) is involved in migration and invasion of keloid fibroblasts (KFs). (a) Differential mRNA expression levels of transforming growth factor-beta 1 (TGF-β1) and TXNDC5 in normal fibroblasts (NFs) and KFs. (b) Differential protein expression levels of TGF-β1 and TXNDC5 in NFs and KFs. (c) Differential mRNA expression levels of Smad2 and Smad3 in NFs and KFs. (d) Reverse transcription-quantitative polymerase chain reaction was used to determine transfection efficiency of si-TXNDC5 in KFs. (e and f) The protein and messenger RNA level of TGF-β1 in KFs treated with si-TXNDC5. (g) Evaluation of fibroblast invasiveness using the Transwell invasion assay. (h) Assessment of fibroblast migration at 48 h in a wound healing assay. Where *P < 0.05, **P < 0.01,***P < 0.001, and ****P < 0.0001.
DISCUSSION
Deciphering the cellular components of keloid is hindered by its intricate microenvironment that consists of numerous fibroblasts, endothelial cells, immune cells, and ECM deposits.[28] Understanding the pathogenic mechanisms of keloid is challenging due to the complex cellular composition and intercellular communication of the dermal disorder.[29] Furthermore, the specific cellular constituents are also affected by transcriptomic and epigenetic modifications.[1,29] As a cutting-edge technique, scRNAseq overcomes the limitations of conventional sequencing methods by enabling the discrimination of distinct cell types within intricate populations.[30-32] Advanced scRNA-seq technology facilitates the profiling of the cellular components of keloids and allows for recording transcriptomic information from individual cells and clustering cells in a vast dataset.[33] Here, a single-cell atlas of keloid was constructed and explored the characteristics and key regulatory pathways of different fibroblast subpopulations were explored. The effect of TXNDC5, a gene associated with fibrogenesis, on keloid was also explored. These findings contributed to a better understanding on the pathogenesis of keloid and provided potential targets for clinical treatment of patients with keloid.
Fibroblasts have been widely studied in various fibrotic diseases.[16] Here, five cell types for keloid based on the scRNA-seq data in GSE163973 was identified and, further, clustered fibroblasts into two fibroblast subpopulations (fibroblast cells 1 and fibroblast cells 2). Enrichment analysis demonstrated that fibroblast subpopulations were mainly involved in processes related to the Wnt signaling pathway, epithelial cell proliferation, the MARK signaling pathway, etc. Among them, the Wnt signaling pathway target genes have been shown to regulate a number of benign pathological changes in the skin.[34,35] Researchers have found aberrant activation of the Wnt/b-catenin signaling pathway in keloid.[36] Furthermore, Liu et al. observed that by lower RNAPTPN12 expression could promote KF growth by targeting micro RNAs to activate the Wnt signaling pathway.[37] Solé-Boldo et al. performed a single-cell analysis based on normal skin samples and classified fibroblasts using unsupervised clustering, and they found that certain fibroblast clusters were enriched in pathways related to inflammation, cellular chemotaxis, ossification, and collagen synthesis.[23] Similarly, previous study classified fibroblasts in keloid into seven subpopulations and found two major KF subpopulations involved in ossification, skeletal system development, and ECM proteoglycan-associated pathways, suggesting the mesenchymal characteristics of these subpopulations.[38] Notably, some genes with similar functions were high-expressed in the two fibroblast subpopulations that identified. For example, COL1A1, COL1A2, and COL3A1 are three isoforms of collagen, which is one of the major components in the tissues such as skin and bone.[39] These genes have been shown to provide mechanical strength and stability to tissues and to promote cell migration and proliferation during the wound healing process to accelerate wound repair.[40-42] SFRP2 promotes the proliferation of cardiac fibroblasts through the activation of the Wnt/b-catenin pathway and is also involved in the regulation of ECM synthesis and degradation as well as biological processes such as cell growth, differentiation, and migration.[43] Kobayashi et al. found that SFRP2-knockout mice have reduced myocardial fibrosis after myocardial infarction as compared to normal mice.[44] These results suggested that the two fibroblast subpopulations may play an important role in keloid and could serve as novel targets for fibrosis treatment.
Intriguingly, both the fibroblast subpopulations were primarily involved in transforming TGF-β signaling pathway.[45] TGF-β is a multifunctional family of cytokines, and the importance of well-regulated TGF-β signaling in wound healing has been reported in the treatment of patients with keloid pathology and chronic injuries.[46] Wang et al. indicated that in keloid with proliferative properties, TGF-β is high-expressed in the tissue and cultured fibroblasts.[47] TXNDC5 was significantly positively correlated with fibroblast genes in the proliferation, migration, and TGF-β signaling pathways. Furthermore, cellular experiments confirmed that the expressions of TXNDC5, TGF-β1, Smad2, and Smad3 were significantly upregulated in KFs. This revealed that TXNDC5 was involved in the proliferation and migration of fibroblasts in keloid through the TGF-β signaling pathway. TXNDC5, an essential part of the PDI family, serves as a chaperone in the ER.[13] Then, TXNDC5 interacts with numerous cellular proteins, facilitating their appropriate folding and ensuring an accurate creation of disulfide bonds at its thioredoxin domains.[48,49] A study with renal fibrosis showed that TXNDC5 is significantly upregulated in fibrotic liver tissues of humans and mice, and that TGF-β1 induces TXNDC5 production in human hepatic stellate cells, which is necessary for the cell activation, proliferation, and survival as well as the production of ECM.[50] Mechanistically, TGF-β1 induces TXNDC5 expression by increasing ER stress and ATF6-mediated transcriptional regulation.[50] Lee et al. found that TXNDC5 expression is upregulated in fibroblasts in pulmonary fibrosis and lungs.[11] TXNDC5 binds directly to TGF-β receptor type 1 (TGFbR1) in lung fibroblasts and enhances TGF-β1 signaling, which, in turn, leads to hyperactivation of fibroblasts, proliferation, and ECM production.[11] Overall, TXNDC5 is associated with fibrosis in a variety of human organs. Therefore, it is reasonable to assume that TGF-β1 combined with TGFbR to activate ER stress and transcription factor ATF6 and, then, upregulated the expression of TXNDC5, which, further, enhanced the pro-fibrotic TGF-β signal through increasing the folding and stability of TGFbR1 protein to promote fibrosis, ultimately leading to keloid.
However, there were some limitations to our study. First, the sample size of keloid in the GEO dataset was relatively small. Second, there was heterogeneity among keloid patients; therefore, the generalizability of the current results should also be validated with larger sample size. Third, the role of TXNDC5 in keloid formation needed to be verified using in vivo animal model and clinical samples. To improve the reliability of our findings, clinical data (database or hospital) were now collected to further validate the conclusions.
SUMMARY
Results confirmed that fibroblasts in keloid were heterogeneous based on the scRNA-seq dataset. High-expressed genes in different fibroblast subpopulations were identified to be able to specifically characterize the function of fibroblast subpopulations. Genes involved in cell proliferation, migration, and TGF-β signaling pathway were more strongly expressed in the fibroblast cells 1 subpopulation. In particular, TXNDC5 was significantly positively correlated with fibroblasts in keloid in terms of proliferation, migration and TGF-β signaling pathway. Those results provided a new direction for the treatment and diagnosis of keloid patients.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
ABBREVIATIONS
TXNDC5: Thioredoxin domain-containing protein 5
GSEA: Gene Set Enrichment Analysis
KFs: keloid fibroblasts
TGF-β: transforming growth factor beta
ECM: extracellular matrix
PDI: protein disulfide isomerase
ER: endoplasmic reticulum
JNK: N-terminal kinase
ERK: extracellular signal-regulated kinase
scRNA-seq: single-cell RNA sequencing
GEO: gene expression omnibus
UMAP: uniform manifold approximation and projection
DMEM: Dulbecco’s modified Eagle medium
Si: small interfering
RT-qPCR: Reverse transcription-quantitative polymerase chain reaction
RIPA: radioimmunoprecipitation assay
BCA: bicinchoninic acid
TBST: Tris-buffered saline with Tween-20
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
PBS: Phosphate-Buffered Saline
mRNA: messenger RNA
ATCC: American Type Culture
AUTHOR CONTRIBUTIONS
ZKL: Concepted and designed the research; LNX and JML: Acquired the data; SDZ and HJX: Analyzed and interpreted data; ZKL, JML and LNX: Drafted the manuscript; SDZ, ZKL and HJX: Revised manuscript for important intellectual content. All authors contributed to this present work. All authors read and approved the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This research did not involve human or animal studies, and therefore ethics committee approval is not required.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
Supplementary material associated with this article can be found:
FUNDING
Not Applicable.
References
- The keloid disorder: Heterogeneity, histopathology, mechanisms and models. Front Cell Dev Biol. 2020;8:360.
- [CrossRef] [Google Scholar]
- Trichostatin A inhibits collagen synthesis and induces apoptosis in keloid fibroblasts. Arch Dermatol Res. 2011;303:573-80.
- [CrossRef] [Google Scholar]
- Molecular signalings in keloid disease and current therapeutic approaches from natural based compounds. Pharm Biol. 2015;53:457-63.
- [CrossRef] [Google Scholar]
- Keloids and scars: A review of keloids and scars, their pathogenesis, risk factors, and management. Curr Opin Pediatr. 2006;18:396-402.
- [CrossRef] [Google Scholar]
- miR-194-5p serves a suppressive role in human keloid fibroblasts via targeting NR2F2. Mol Med Rep. 2021;23:57.
- [CrossRef] [Google Scholar]
- Keloids and hypertrophic scars: Pathophysiology, classification, and treatment. Dermatol Surg. 2017;43(Suppl 1):S3-18.
- [CrossRef] [Google Scholar]
- EndoPDI, a novel protein-disulfide isomerase-like protein that is preferentially expressed in endothelial cells acts as a stress survival factor. J Biol Chem. 2003;278:47079-88.
- [CrossRef] [Google Scholar]
- The role and mechanism of TXNDC5 in diseases. Eur J Med Res. 2022;27:145.
- [CrossRef] [Google Scholar]
- TXNDC5 protects synovial fibroblasts of rheumatoid arthritis from the detrimental effects of endoplasmic reticulum stress. Intractable Rare Dis Res. 2020;9:23-9.
- [CrossRef] [Google Scholar]
- Fibroblast-enriched endoplasmic reticulum protein TXNDC5 promotes pulmonary fibrosis by augmenting TGFß signaling through TGFBR1 stabilization. Nat Commun. 2020;11:4254.
- [CrossRef] [Google Scholar]
- Endoplasmic reticulum protein TXNDC5 promotes renal fibrosis by enforcing TGF-ß signaling in kidney fibroblasts. J Clin Invest. 2021;131:143645.
- [CrossRef] [Google Scholar]
- Knockdown of TXNDC5 alleviates CCL4-induced hepatic fibrosis in mice by enhancing endoplasmic reticulum stress. Am J Med Sci. 2023;366:449-57.
- [CrossRef] [Google Scholar]
- The novel role of ER protein TXNDC5 in the pathogenesis of organ fibrosis: Mechanistic insights and therapeutic implications. J Biomed Sci. 2022;29:63.
- [CrossRef] [Google Scholar]
- Keloids: The paradigm of skin fibrosis-Pathomechanisms and treatment. Matrix Biol. 2016;51:37-46.
- [CrossRef] [Google Scholar]
- Understanding the impact of fibroblast heterogeneity on skin fibrosis. Dis Model Mech. 2020;13:10.1242/dmm.044164.
- [CrossRef] [Google Scholar]
- Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science (NY). 2015;348:aaa2151.
- [CrossRef] [Google Scholar]
- Periostin in the pathogenesis of skin diseases. Cell Mol Life Sci. 2017;74:4321-8.
- [CrossRef] [Google Scholar]
- The role of periostin in lung fibrosis and airway remodeling. Cell Mol Life Sci. 2017;74:4305-14.
- [CrossRef] [Google Scholar]
- Fibroblast heterogeneity: Implications for human disease. J Clin Invest. 2018;128:26-35.
- [CrossRef] [Google Scholar]
- Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277-81.
- [CrossRef] [Google Scholar]
- SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J Invest Dermatol. 2018;138:802-10.
- [CrossRef] [Google Scholar]
- Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol. 2020;3:188.
- [CrossRef] [Google Scholar]
- Comprehensive integration of single-cell data. Cell. 2019;177:1888-902.e21.
- [CrossRef] [Google Scholar]
- clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-7.
- [CrossRef] [Google Scholar]
- SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083-6.
- [CrossRef] [Google Scholar]
- An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3:71-85.
- [Google Scholar]
- Galectin-1 and Galectin-3 and their potential binding partners in the dermal thickening of keloid tissues. Am J Dermatopathol. 2019;41:193-204.
- [CrossRef] [Google Scholar]
- Decoding the molecular landscape of keloids: New insights from single-cell transcriptomics. Burns Trauma. 2023;11:tkad017.
- [CrossRef] [Google Scholar]
- Single-cell RNA sequencing reveals potential for endothelial-tomesenchymal transition in tetralogy of Fallot. Congen Heart Dis. 2023;18:611-25.
- [CrossRef] [Google Scholar]
- The progressive application of single-cell RNA sequencing technology in cardiovascular diseases. Biomed Pharmacother. 2022;154:113604.
- [CrossRef] [Google Scholar]
- Single cell RNA sequencing-a valuable tool for cancer immunotherapy: A mini review. Oncologie. 2023;25:635-9.
- [CrossRef] [Google Scholar]
- Single-cell RNA-seq reveals the communications between extracellular matrix-related components and Schwann cells contributing to the earlobe keloid formation. Front Med (Lausanne). 2022;9:1000324.
- [CrossRef] [Google Scholar]
- Upregulation of the Wnt/beta-catenin pathway induced by transforming growth factor-beta in hypertrophic scars and keloids. Acta Derm Venereol. 2006;86:300-7.
- [CrossRef] [Google Scholar]
- Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol. 2008;128:1298-310.
- [CrossRef] [Google Scholar]
- Decoy Wnt receptor (sLRP6E1E2)-expressing adenovirus induces anti-fibrotic effect via inhibition of Wnt and TGF-ß signaling. Sci Rep. 2017;7:15070.
- [CrossRef] [Google Scholar]
- Silencing circular RNAPTPN12 promoted the growth of keloid fibroblasts by activating Wnt signaling pathway via targeting microRNA-21-5p. Bioengineered. 2022;13:3503-15.
- [CrossRef] [Google Scholar]
- Integrated analysis of single-cell and spatial transcriptomics in keloids: Highlights on fibrovascular interactions in keloid pathogenesis. J Invest Dermatol. 2022;142:2128-39.e11.
- [CrossRef] [Google Scholar]
- Specifications and validation of the ACMG/ AMP criteria for clinical interpretation of sequence variants in collagen genes associated with joint hypermobility. Hum Genet. 2023;142:785-808.
- [CrossRef] [Google Scholar]
- COL1A1: A novel oncogenic gene and therapeutic target in malignancies. Pathol Res Pract. 2022;236:154013.
- [CrossRef] [Google Scholar]
- Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med (Wars). 2022;17:329-40.
- [CrossRef] [Google Scholar]
- Velvet antler peptides reduce scarring via inhibiting the TGF-ß signaling pathway during wound healing. Front Med (Lausanne). 2021;8:799789.
- [CrossRef] [Google Scholar]
- sFRP2 activates Wnt/ß-catenin signaling in cardiac fibroblasts: Differential roles in cell growth, energy metabolism, and extracellular matrix remodeling. Am J Physiol Cell Physiol. 2016;311:C710-9.
- [CrossRef] [Google Scholar]
- Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat Cell Biol. 2009;11:46-55.
- [CrossRef] [Google Scholar]
- Current potential therapeutic strategies targeting the TGF-ß/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed Pharmacother. 2020;129:110287.
- [CrossRef] [Google Scholar]
- Transforming growth factor beta (TGF-ß) isoforms in wound healing and fibrosis. Wound Repair Regen. 2016;24:215-22.
- [CrossRef] [Google Scholar]
- Hypertrophic scar tissues and fibroblasts produce more transforming growth factor-beta1 mRNA and protein than normal skin and cells. Wound Repair Regen. 2000;8:128-37.
- [CrossRef] [Google Scholar]
- TXNDC5, a newly discovered disulfide isomerase with a key role in cell physiology and pathology. Int J Mol Sci. 2014;15:23501-18.
- [CrossRef] [Google Scholar]
- Inhibition of TXNDC5 attenuates lipopolysaccharide-induced septic shock by altering inflammatory responses. Lab Invest. 2022;102:422-31.
- [CrossRef] [Google Scholar]
- Targeting ER protein TXNDC5 in hepatic stellate cell mitigates liver fibrosis by repressing non-canonical TGFß signalling. Gut. 2022;71:1876-91.
- [CrossRef] [Google Scholar]








