Translate this page into:
Sterol regulatory element binding transcription factor 1 is an important prognostic factor for colon adenocarcinoma and closely related to immune infiltration

*Corresponding author: Aihua Jin, Ph.D. Central Laboratory, The Affiliated Hospital of Yanbian University, Yanji, Jilin, China. drjinah@163.com
-
Received: ,
Accepted: ,
How to cite this article: Jin L, Lin Z, Jin A. Sterol regulatory element binding transcription factor 1 is an important prognostic factor for colon adenocarcinoma and closely related to immune infiltration. CytoJournal. 2024;21:67. doi: 10.25259/Cytojournal_43_2024
Abstract
Objective:
Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) encodes a core protein that has a crucial function in the metabolism of cholesterol and lipids. This transcription factor is a member of the family of transcription factors and highly expressed in a variety of cancer types. As of now, there are few reports on the relationship between the expression of SREBF1 and colon adenocarcinoma (COAD). Hence, this study utilizes databases and a range of experiments to explore the relationship between the expression of SREBF1 and tumor immune infiltration, as well as the occurrence and development of tumors.
Material and Methods:
The expression of SREBF1 in pan-cancers was retrieved through databases such as TIMER, Gene Expression Profiling Interactive Analysis (GEPIA), and UALCAN. The expression of SREBF1 in HCT-116 and SW480 cells was detected using western blot. Furthermore, we also found that knockdown SREBF1 can inhibit the proliferation and migration of COAD cells. The correlation between SREBF1 and autophagy in COAD cells was detected using acridine orange (AO) staining, western blot, and immunofluorescence (IF).
Results:
The databases of TIMER, GEPIA and UALCAN revealed that SREBF1 is overexpressed in pan-cancer tissues, and closely associated with the prognosis of the patients with cancer. Further immunohistochemical staining showed that SREBF1 was overexpressed in COAD, and closely related to the clinical stage and lymph node metastasis. Western blot revealed that SREBF1 was significantly expressed in both HCT-116 and SW480 COAD cells; knockdown of SREBF1 could inhibit the proliferation, DNA replication, and migration of COAD cells. The AO staining, western blot, and IF experiments also showed that silencing SREBF1 could promote the autophagy of COAD cell. Meanwhile, the TIMER database indicates a significant positive correlation between the presence of immune cells in COAD and variations in copy number alteration of SREBF1.
Conclusion:
SREBF1 might serve as a potential prognostic marker for COAD and be associated with immune cell infiltration.
Keywords
Sterol regulatory element binding protein 1
Colonic neoplasms
Cell proliferation
Transcellular cell migration
Prognosis
INTRODUCTION
The most common cause of death from cancer is colon adenocarcinoma (COAD), which is also one of the most common types of cancers seen all over the globe.[1] It exhibits a high incidence rate, elevated mortality, and an unfavorable prognosis.[2] At present, the therapeutic modalities for COAD typically encompass surgical intervention, radiotherapy, and chemotherapy.[3-5] COAD occurrence is related to various factors including age, family history, and individual genetic factors.[6] Early diagnosis is crucial for COAD treatment and prognosis. However, patients with COAD are usually diagnosed at an advanced stage.[7] Therefore, further research on effective biomarkers for the early COAD detection is necessary. A comprehensive grasp of the molecular mechanisms underlying COAD can enhance patient quality of life.
Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) is a crucial factor in lipid production pathways.[8] It is abnormally expressed in various cancers and provides biosynthetic substances necessary for cancer cell proliferation and survival.[9] Tan et al. showed that the SREBF1 activity is related with head-and-neck squamous cell carcinoma (HNSC) cell proliferation and migration. SREBF1 knockdown may inhibit the abnormal proliferation of HNSC cells, thereby supporting tumor growth.[10] Wang et al. found that the SREBF1 axis facilitated the progression and metastasis of hepatocellular carcinoma by regulating the stability of SNAIL.[11] However, the relevant SREBF1 mechanisms in COAD development are not yet clear, and further, in-depth research is needed.
Tumor microenvironment (TME), which is an intricate amalgamation of several types of cells, blood vessels, extracellular matrix, and solutes surrounding a tumor, significantly impacts tumor development, growth, and therapeutic response.[12,13] Immune infiltration, which refers to immune cell presence and activity in the tumor tissues, is an important aspect closely related to the TME. Exploring the correlation between immune cell infiltration and TME can also provide new guidance for tumor treatment defense.[14] By modulating immune cell functions within the TME, the immune system can enhance its response to tumors and improve the immunotherapy success rate.[15] Therefore, identifying new immunotherapeutic targets could provide new guidance for treating patients with COAD in clinical practice.
SREBF1 expression, gene mutations, and immune infiltration were all explored in this research, which was conducted on a number of different malignancies, particularly through cell experiments, to verify whether SREBF1 promotes COAD cell proliferation and migration in vitro, which are closely related to autophagy. The results indicate that SREBF1 might serve as a potential biomarker, providing novel insights for the prognosis and treatment of patients with COAD.
MATERIAL AND METHODS
Clinical specimens
The tissue microarrays were purchased from Outdo Biotech Co., Ltd. (Shanghai, China), including COAD with clinical features (94 cases) and adjacent tissues (86 cases), and signed informed consent forms before collection. The 8th edition guidelines from the Joint Committee on Cancer Staging in the United States are the basis for the assessment and classification.[16]
Cell lines and reagents
The COAD cell lines include HCT-116 (ATCC®Number: CCL-247™; Organism: Homo sapiens [human]), SW480 (ATCC®Number: CCL-228™; Organism: Homo sapiens [human]), SW620 (ATCC®Number: CCL-227™; Organism: Homo sapiens [human]), and HT-29 (ATCC®Number: HTB-38™; Organism: Homo sapiens [human]) from Zhibei Biotechnology Co., Ltd. (Shanghai, China), and all above cells were identified by short tandem repeat and detected by mycoplasma. SREBF1 short interfering RNA (siRNA) (si-Control, si-SREBF1#1, si-SREBF1#2, and si-SREBF1#3) was purchased from RiboBio (Guangzhou, China). The knockdown effects of si-Control, si-SREBF1#2, and si-SREBF1#3 were significant, and further research was conducted. Following the manufacturer’s instructions to the letter, Lipofectamine 3000 (L3000015, ThermoFisher, USA) was used to transfect the HCT-116 and SW480 cell lines with 30 nM siRNA. Technical information on antibodies, reagents, and instrumentation is provided in Supplementary Tables 1-3.
Immunohistochemistry (IHC)
Take out the tissue microarray in advance and let it sit at room temperature for 10 min, then heat it in an oven for 1 h. Soak with xylene dewax, alcohol, etc. Perform antigen repair in heated sodium citrate. Add 3% peroxidase gradually over 20 min. Administer the primary antibody and let it incubate overnight at 4°C. On the 2nd day, after cleaning, secondary antibody treatment was performed for 1 h, and stain with 3,3’-diaminobenzidine, and finally score all samples based on staining intensity and area.
SREBF1, which localized in a brownish hue within the cytoplasm or nucleus, was deemed positive. Assessment relied on both the staining intensity of positive cells and the proportion of positively stained cells. (1) Evaluate based on the staining intensity of positively identified cells: 0 for no staining; 1 for light yellow; 2 for brown; 3 for yellowish brown. (2) Score according to the percentage of positive cells: 0–10% positive cells is 0; 11–25% is 1; 26–50% is 2; 51–75% is 3; and >75% is 4. The results of the above two scores were multiplied: 0–3 as (-), 4–6 as (+), 7–9 as (++), and 10–12 as (+++). Among them, (++) and (+++) are defined as high SREBF1 expression.
Western blot
Proteins were isolated from cells utilizing Radio Immunoprecipitation Assay Lysis Buffer and quantified using the Bicinchoninic acid protein quantification kit (CW0014S, Cwbio, China) to determine the extracted protein concentrations at 3.0 µg/µL. Polyacrylamide gel electrophoresis with sodium dodecyl sulfate was used for protein separation. The samples subsequently underwent transfer onto a Polyvinylidene fluoride (PVDF) membrane (IPVH00010, Merck Millipore Ltd, Ireland) utilizing semi-dry transfer apparatus (BioRad, USA). The PVDF membrane was submerged in milk (232100, BD company, USA) for 1 h. The primary antibody was added to bind to the target protein. The following day, after washing with Tris Buffered Saline with Tween-20, secondary antibodies were added and detected using chemiluminescence. Finally, statistical analysis was conducted using ImageJ software (National Institutes of Health. Java 1.8.0 345 (64-bit).
Cell counting kit-8 (CCK8) assay
The cells were cultivated in a 96-well plate, maintaining a density of 2000 cells per well. Reagents for siRNA transfection were given to the cells, and the rate of cell proliferation was measured at 0, 24, 48, and 72 h after its administration. The CCK-8 reagent (PF00004, Proteintech, China) and culture medium were combined in a 1:1 ratio to create a CCK-8 working solution. The cells were then incubated with this solution for 1–4 h. Absorbance was measured at 595 nm. Finally, the data underwent analysis using GraphPad Prism 9.0 software.
Colony formation assay
COAD cells were grown to approximately 90% confluence for digestion. A cell counting plate (Countstar, USA) was used to count 1000 cells per well, and the cells were removed after 14 days of cell culture. Subsequently, the samples underwent three washes with phosphate-buffered saline (PBS) and were then fixed using a 4% paraformaldehyde solution and stained. Finally, they were imaged and statistically analyzed.
5-ethynyl-2’-deoxyuridine (EdU) assay
After cell counting, 5000 cells/well were inoculated onto a 96 well plate. Adding siRNA 48 h to COAD cells, add culture medium containing EdU for 2 h follow the instructions for the EdU reagent kit (C10310-1, RiboBio, China). The cells underwent a washing process using PBS and underwent fixation with 4% paraformaldehyde. The cell membrane was perforated with 0.1% Triton X-100 solution and the pre-prepared ×1 Hoechst 33342 staining solution was added. Subsequently, the stained cells underwent observation through fluorescence microscopy. The results were analyzed using ImageJ software.
Wound healing assay
After cell counting, 3 × 105 cells per well were inoculated into 96-well plates. A wound was created on the cell monolayer using the cell scraping method. After scratching, cell fragments were removed and washed. Immediately after the wound was formed, microscopic images of the wound area were captured to record the wound conditions at 0 h. The cells were cultured and changes in the wound area were regularly observed for 48 h. Automatic image analysis tools were used to quantitatively analyze the wound area width.
Transwell assay
Introduce 3 × 105 cells into each well of the small chamber. Subsequently, add 1 mL of culture media containing 20% fetal bovine serum (A5670701, GIBCO, USA) to the lower chamber. After 20–24 h of observation, the cells were washed with PBS after passing through the chamber, and 4% paraformaldehyde was used for cell fixation. Finally, stain with hematoxylin (H8070, Solarbio, China). Seal the small chamber membrane on a glass slide with neutral resin. Observe it under an optical inverted microscope (Olympus, Japan). Then, conduct image acquisition for counting and statistical analysis.
Acridine orange (AO) staining
After treating COAD cells with siRNA for 48 h, they were stained with 1 µg/mL AO (A6009, Macklin, China) and washed with PBS in the dark. Finally, acidic vesicle organelle formation (bright red fluorescence in acidic vesicles) was measured using fluorescence microscopy.
Immunofluorescence (IF) staining
Following a 48 h incubation period, proceeded to fix the cells using 4% paraformaldehyde. Subsequently, the cell membrane underwent permeabilization through the application of 0.1% Triton X-100 and a combination of other detergents. The cells underwent overnight incubation at 4°C with a suitable quantity of monoclonal antibody. The unbound antibodies were washed using buffer solutions such as PBS. A fluorescent secondary antibody bound to the primary antibody was then added. The target proteins were ultimately examined using a fluorescence upright microscope (Olympus, Japan).
Bioinformatic analysis
The databases of Tumor Immune Estimation Resource (TIMER) (https://cistrome.shinyapps.io/timer/),[17] The University of Alabama at Birmingham Department of Pathology (UALCAN) (http://uaLcan.path.uab.edu),[18] Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA/#/)[19] and Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn)[20] were used to explore SREBF1 expression in pan-cancer tissues and the relationship between SREBF1 overexpression and clinical features of the patients with cancer, such as survival, differentiation, and cancer stage. The cBioportal database (http://www.cbioportal.org/)[21] contains large-scale cancer genomic data and provides information on SREBF1 mutation frequency, copy number alteration (CNA), and mutation type. Sangerbox (http://sangerbox.com/home.html)[22] found a relationship between the expression of SREBF1 relating to patient microsatellite instability (MSI) and tumor mutation burden (TMB), Neoantigen, DNAss, RNAss, and differentially methylated probes-based (DMPss) in pan-cancer, especially in COAD. The STRING database (https://string-db.org/)[23] was utilized to determine the proteins that associate with SREBF1. The findings demonstrated a relationship between SREBF1 expression levels and the infiltration of immune cells in patients with COAD, as evidenced by the analyses from TIMER and GSCA.
State ethics and informed consent
The Declaration of Helsinki’s ethical guidelines were followed in the conduct of this investigation.[24] The Shanghai Outdo Biotech Company’s Institutional Review Board (IRB) examined and approved the research procedure. The IRB approval number for this study is YB M-05-02. All participants, as well as their legal guardians, have submitted signed informed consent forms. Participants are assured that their participation is voluntary. The consent form and process have also been approved by IRB.
Statistical analysis
The data were analyzed with GraphPad Prism 9.0 software. Paired t-tests of independent means were used for intergroup comparisons. The survival curve was generated using Kaplan– Meier analysis. The continuous data were categorized and compared using one-way analysis of variance, and each experiment was performed three times. A significance threshold of P < 0.05 was used to ascertain statistical significance.
RESULTS
SREBF1 expression in pan-cancer tissues
The TIMER database showed SREBF1 overexpression in most cancer tissues than that in normal tissues, including bladder urothelial carcinoma (BLCA), lung adenocarcinoma (LUAD), breast invasive carcinoma (BRCA), COAD, kidney chromophobe, esophageal carcinoma, HNSC, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma, prostate adenocarcinoma, lung squamous cell carcinoma, rectum adenocarcinoma (READ), thyroid cancer (THCA), and uterine corpus endometrial carcinoma (UCEC) [Figure 1a]. Moreover, the UALCAN databases provided further confirmation of SREBF1 overexpression in tumor tissues compared to the normal tissues across different types of malignancies [Figure 1b]. In sum, these data indicate that SREBF1 is significantly upregulated in many cancer types.
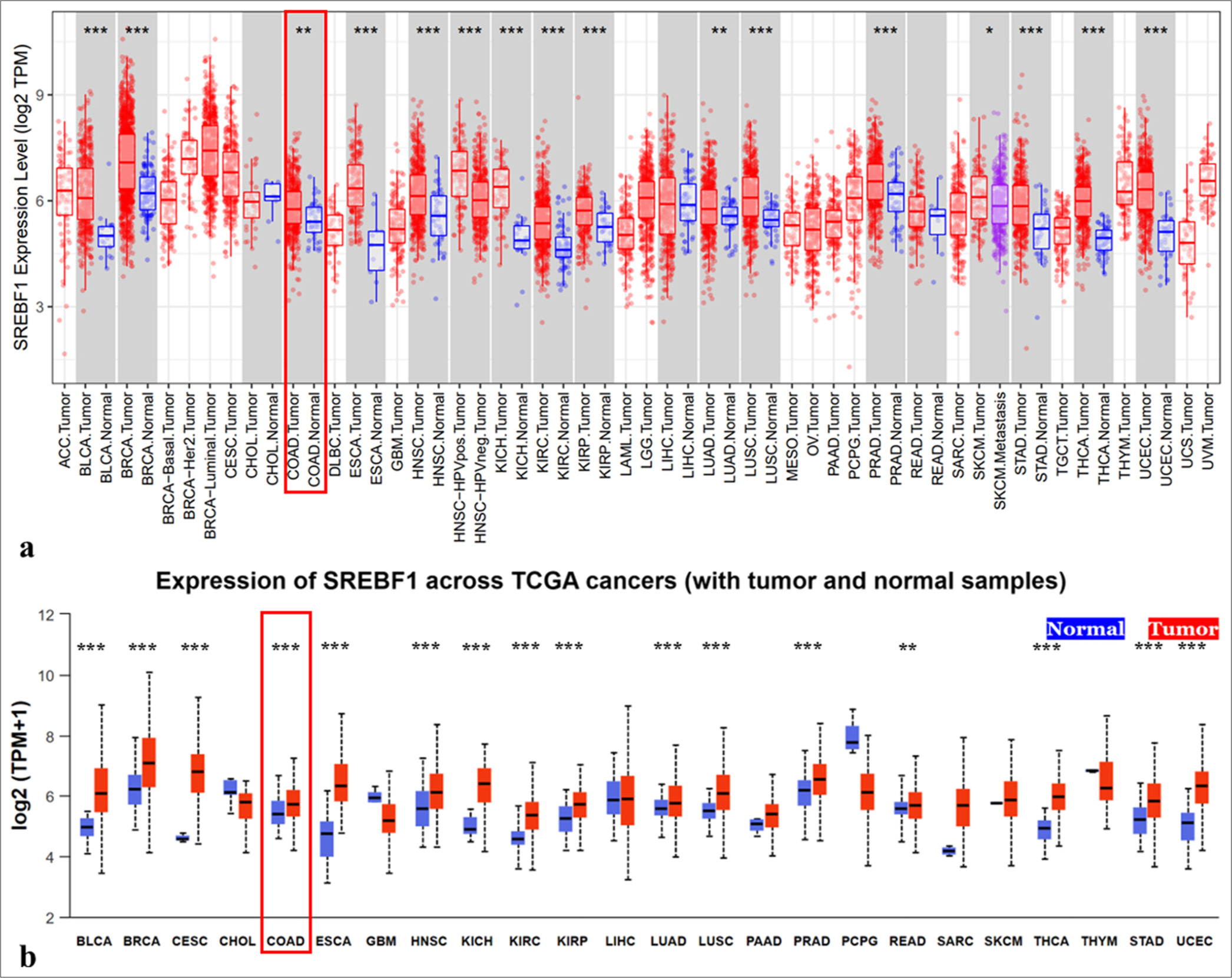
- The expression of Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) in pan-cancer tissues. (a and b) Expression of SREBF1 in pan-cancers from the TIMER and UALCAN databases (✶P < 0.05, ✶✶P < 0.01, ✶✶✶P < 0.001). TCGA: The Cancer Genome Atlas Program, ACC: Adrenocortical carcinoma, BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and neck squamous cell carcinoma, HPV: Human Papillomavirus, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin Cutaneous Melanoma, STAD: Stomach adenocarcinoma, TGCT: Testicular Germ Cell Tumors, THCA: Thyroid cancer, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma, UCS: Uterine Carcinosarcoma, UVM:Uveal Melanoma.
Overexpression of SREBF1 is strongly associated with a poor prognosis in the patients
To get a deeper understanding of the expression of SREBF1 in COAD, further IHC was done. And the results showed that SREBF1 expression was significantly up-regulated in COAD tissues compared to the normal counterpart [Figure 2a]. Fifty out of 86 normal tissues showed positive SREBF1 expression, with a positive rate of 58.1%, while 85 out of 94 COAD tissues exhibited positive SREBF1 expression, with a much higher positive rate of 90.4% (P < 0.05). Furthermore, the rate of SREBF1 expression in COAD tissues was strongly positive at 65.0%, which was significantly greater than the rate in non-tumor tissues (19.8%, P < 0.05) [Table 1]. Meanwhile, a positive clinicopathological correlation was found between SREBF1 expression and clinical stage (P = 0.003) as well as lymph node metastasis (P = 0.026), as shown in Table 2. The preliminary database revealed that SREBF1 is highly expressed in various tumors, and the GSCA database further found that SREBF1 is closely related to the clinical stage of various tumors. For example, it is significantly positively correlated with tumors such as BRCA, CHOL, BLCA, liver hepatocellular carcinoma, LUAD, and THCA. Meanwhile, the expression of SREBF1 had a strong inverse association with the occurrence of many malignancies, including KIRC, testicular germ cell tumors, mesothelioma (MESO), and READ [Figure 2b]. Subsequently, analysis using GEPIA and Kaplan–Meier survival curves has shown a significant association between elevated expression of SREBF1 and an unfavorable prognosis in several cancers, closely related to overall survival of BLCA, acute myeloid leukemia, MESO, COAD, and ovarian cancer (OV), disease-free survival of COAD, progression free survival of OV and patient protected study (PPS) of COAD. It is worth noting that after high expression of SREBF1, the PPS of COAD patients is prolonged, which may be related to multiple factors such as patient sample size and cancer differentiation, further research is needed. In summary, the elevated expression of SREBF1 in many types of malignancies is linked to worse prognosis in patients (P < 0.05, [Figure 2c and d]).
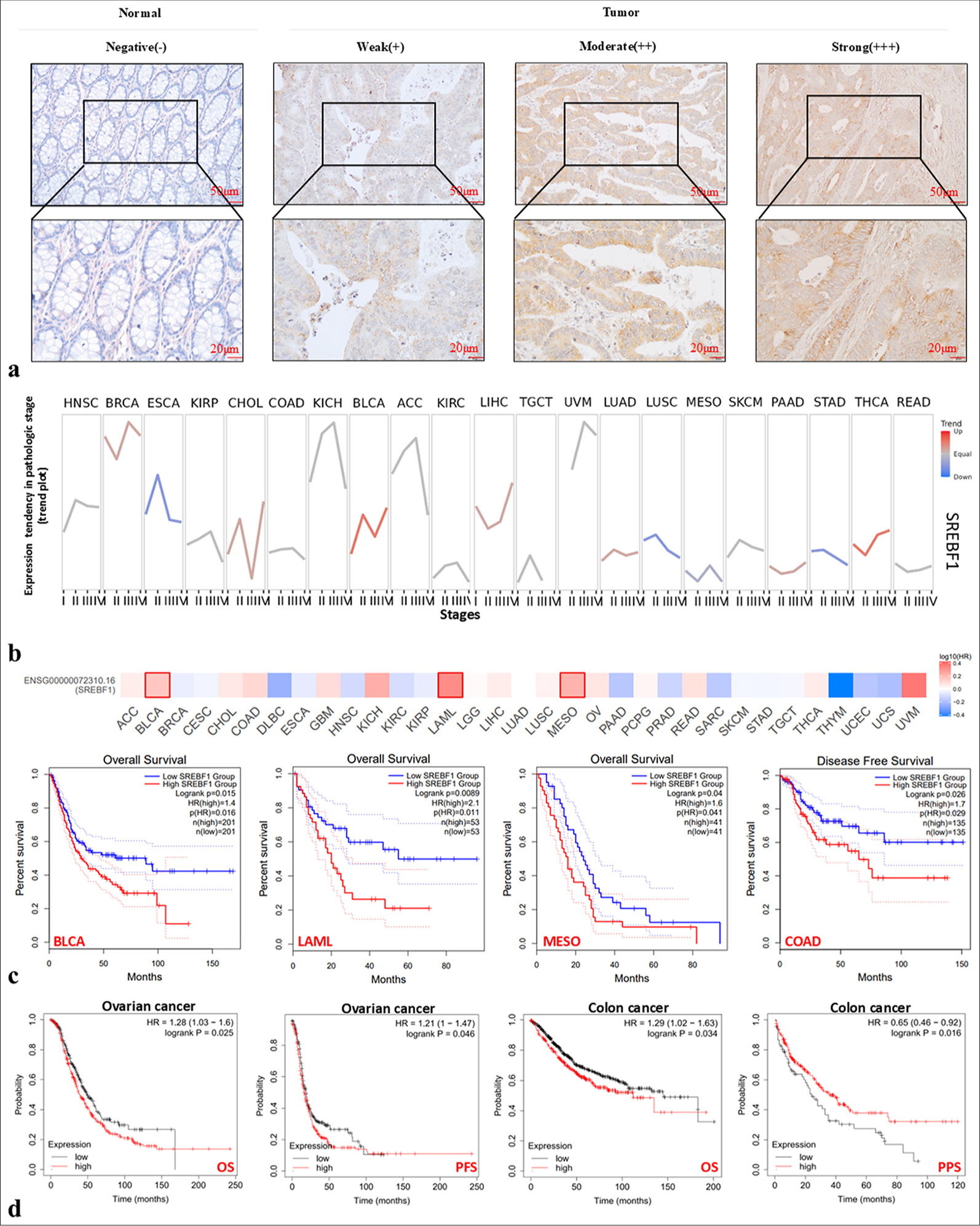
- The high expression of Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) is closely related to the clinical staging of patients and their prognosis. (a) Immunohistochemical staining showed that SREBF1 was greatly overexpressed in colon adenocarcinoma tissues compared to normal tissues. (b) The correlation between SREBF1 expression level and clinical stage in pan-cancer patients via Gene Set Cancer Analysis database. (c and d) Detect the prognostic correlation between SREBF1 expression level and patient survival through the Gene Expression Profiling Interactive Analysis database. ACC: Adrenocortical carcinoma, BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and neck squamous cell carcinoma, HPV: Human Papillomavirus, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin Cutaneous Melanoma, STAD: Stomach adenocarcinoma, TGCT: Testicular Germ Cell Tumors, THCA: Thyroid cancer, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma, UCS: Uterine Carcinosarcoma, UVM: Uveal Melanoma.
| Group | n | Sterol Regulatory Element Binding Transcription Factor 1 protein expression | Positive rate (%) | Strong positive rate (%) | |||
|---|---|---|---|---|---|---|---|
| - | + | ++ | +++ | ||||
| Colon adenocarcinoma | 94 | 9 | 24 | 23 | 38 | 90.4✶✶ | 64.9✶✶ |
| Paracancerous tissue | 86 | 36 | 33 | 9 | 8 | 58.1 | 19.8 |
✶✶The difference is statistically significant, P<0.01. Positive rate: The percentage of samples with staining intensity of +, ++ and +++, Strong positive rate: The percentage of samples with staining intensity of++and +++
| Clinical pathological parameters | n | Sterol regulatory element binding transcription factor 1 strong positive rate (%) | Chi-square value | P-value |
|---|---|---|---|---|
| Gender | 0.004 | 0.949 | ||
| Male | 48 | 31 (64.6) | ||
| Female | 46 | 30 (65.2) | ||
| Age (years) | 0.605 | 0.437 | ||
| ≤65 | 49 | 30 (61.2) | ||
| >65 | 45 | 31 (68.9) | ||
| Tumor size (cm) | 0.657 | 0.418 | ||
| >5 | 43 | 20 (46.5) | ||
| ≤5 | 51 | 28 (54.9) | ||
| Differentiation | 0.014 | 0.907 | ||
| High+medium | 85 | 55 (64.7) | ||
| Low | 9 | 6 (66.7) | ||
| Clinical stage | 8.708 | 0.003✶✶ | ||
| I+II | 58 | 31 (53.4) | ||
| III+IV | 36 | 30 (83.3) | ||
| pT | 1.566 | 0.211 | ||
| T1-2 | 11 | 9 (81.9) | ||
| T3-4 | 83 | 52 (62.7) | ||
| Lymph node metastasis | 4.928 | 0.026✶ | ||
| Yes | 34 | 27 (79.4) | ||
| No | 60 | 34 (56.7) |
✶The difference is statistically significant, P<0.05, ✶✶The difference is statistically significant, P<0.01
Gene mutations and immune markers of SREBF1 in pan-cancer cells
Gene and protein mutations may be important factors in promoting cancer development. According to the search results on the cBioportal website, we found that SREBF1 mutation frequency was very high, mainly amplifications and mutations, particularly in SARC and UCEC [Figure 3a].
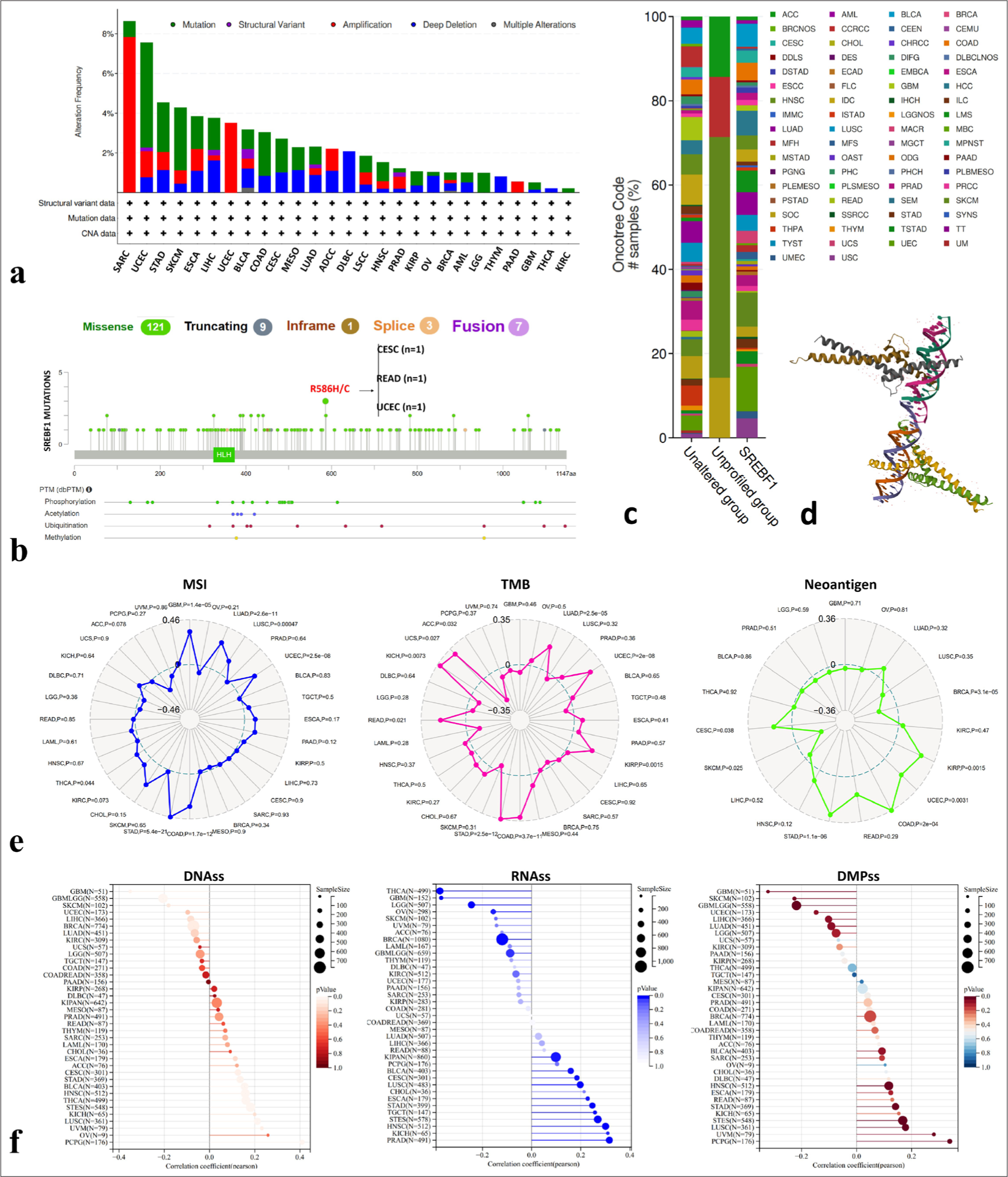
- Mutation characteristics and immunogenicity indicators of the Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) gene in pan-cancer. (a-c) Genetic change types, frequencies, and mutation sites of SREBF1 in The Cancer Genome Atlas Program (TCGA) pan-cancer obtained through cBioportal database. (d) The 3D structure displays a model image of SREBF1 on the cBioportal database. (e) Sanger Box described the correlation analysis between the expression of SREBF1 in pan-cancer cells and microsatellite instability, tumor mutation burden and neoantigen. (f) Sanger Box described the correlation analysis between SREBF1 expression, DNAss, RNAss, and DMPss in pan-cancer. TGCA: The Cancer Genome Atlas Program, ACC: Adrenocortical carcinoma, BLCA: Bladder urothelial carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, DLBC: Lymphoid neoplasm diffuse large B-cell lymphoma, ESCA: Esophageal carcinoma, GBM: Glioblastoma multiforme, HNSC: Head and neck squamous cell carcinoma, HPV: Human papillomavirus, KICH: Kidney chromophobe, KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute myeloid leukemia, LGG: Brain lower grade glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma, MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin cutaneous melanoma, STAD: Stomach adenocarcinoma, TGCT: Testicular Germ Cell Tumors, THCA: Thyroid cancer, THYM: Thymoma, UCEC: Uterine corpus endometrial carcinoma, UCS: Uterine carcinosarcoma, UVM: Uveal melanoma, CNA: Copy number alteration,
Genetic alteration was further detected among amino acids 0 and 1288 of SREBF1, and the missense mutation was regarded as the dominant factor, except for a variant of undetermined significance mutation. R586H/C was the most frequent mutation site in SREBF1 in cervical squamous cell carcinoma and endocervical adenocarcinoma (n = 1), READ (n = 1), and UCEC (n = 1) [Figure 3b]. The OncoTree code showed the frequency of genetic modifications in pan-cancer types [Figure 3c] and the 3D structure of SREBF1 is shown in [Figure 3d]. In addition, we searched for correlations among MSI, TMB, and SREBF1 neoantigens in pan-cancer tissues using the Sanger box [Figure 3e]. Cancer stemness affects the occurrence, metastasis, and drug resistance in various cancers. The relationship between pan-cancer tumor stemness and SREBF1 expression was detected through DNAss, RNAss, and DMPss [Figure 3f]. In sum, we have discovered a correlation between cancer stem cells and the immunological milieu of the tumor, which has implications for treatment options in clinical patients and has significant potential for the future.
Silencing SREBF1 inhibits COAD cell proliferation
The STRING website was used to analyze the genes [Figure 4a], biological processes, molecular functions, protein domains, and pathway enrichment of SREBF1 in COAD and found that SREBF1 affects pathways such as lipid metabolism, cell growth, apoptosis, and immunity [Figure 4b]. To elucidate SREBF1 regulation in the biological function of COAD, we detected SREBF1 protein expression in COAD cell lines (HCT-116, SW480, SW620, and HT-29), and the results showed significantly higher expression of HCT-116 and SW480 relative to SW620 and HT29. Therefore, HCT-116 and SW480 cells were selected for subsequent experiments [Figure 4c and Supplementary Figure 1a]. Subsequently, both HCT-116 and SW480 cells were transfected with non-targeting siRNA. Compared with the si-Control group, both the transfection effects of the si-SREBF1#2 and si-SREBF1#3 groups were more pronounced as shown by the western blot [Figure 4d, Supplementary Figure 1b and c]. CCK8 assay demonstrated that inhibiting SREBF1 could greatly suppress the proliferation of COAD cells [Figure 4e]. EdU assay showed that SREBF1 silencing could significantly inhibit DNA replication in COAD cells [Figure 4f]. Furthermore, in the Colony formation assay, si-SREBF1#2 and si-SREBF1#3 could reduce the colony-forming capacity of COAD cells [Figure 4g]. These studies indicate that silencing SREBF1 may inhibit COAD cell proliferation.
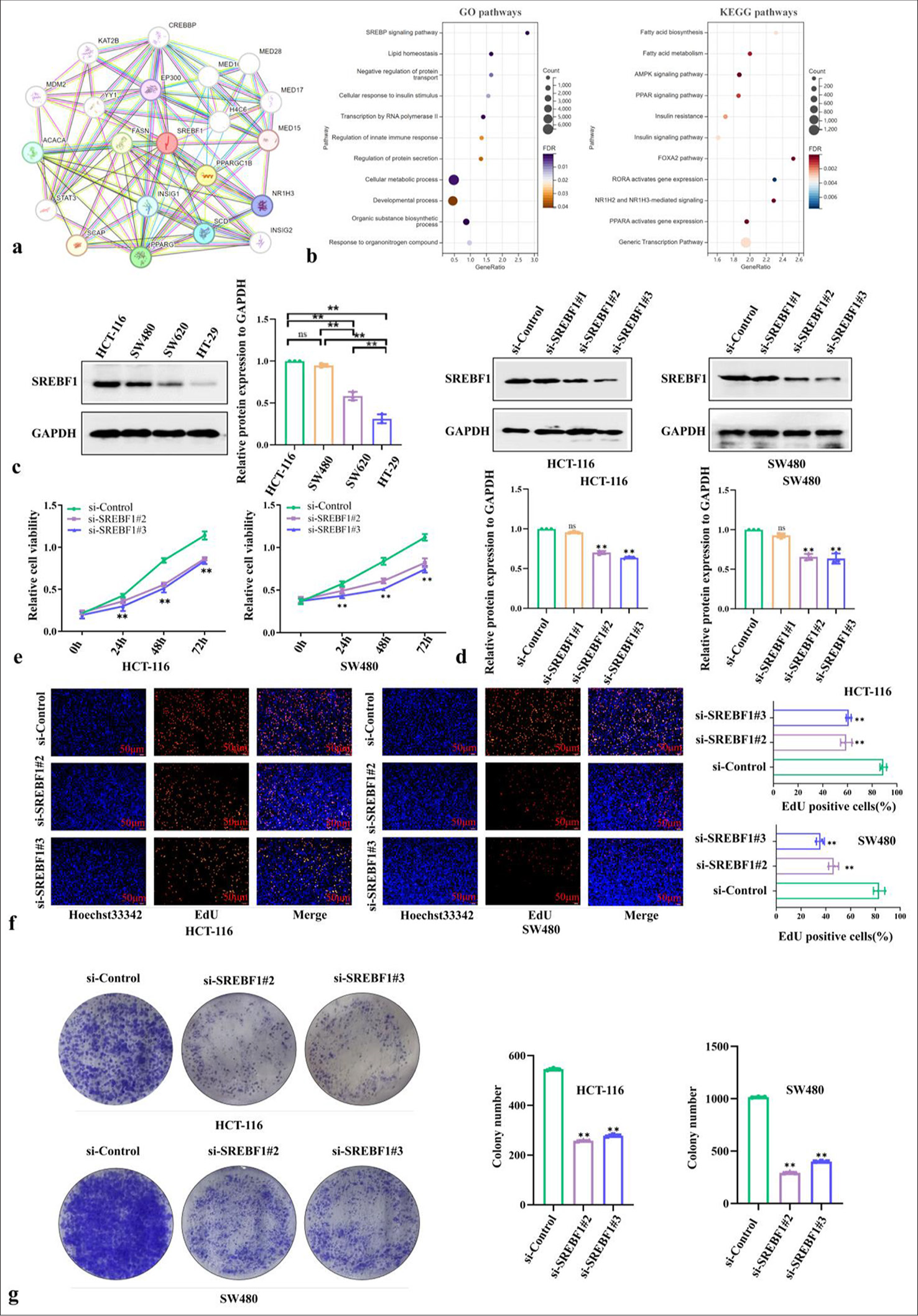
- Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) contributes to proliferation and autophagy related processes of colon adenocarcinoma. (a) The genes related to SREBF1 were identified using STRING (https://cn.string-db.org/). (b) The Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis were also displayed based on the related genes mentioned above. (c) Screening colon adenocarcinoma cells with significant SREBF1 expression through western blot, Glyceralde-hyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (d) Verify the transfection efficacy of different si-SREBF1 sequences through western blot, GAPDH was used as a loading control. (e-g) Cell proliferation was tested by Cell counting kit-8, 5-ethynyl-2’-deoxyuridine (50 µm), and colony formation. Data were shown as mean±standard error of at least three independent experiments (n = 3) (ns: Not significant; ✶P < 0.05; ✶✶P < 0.01). GO: Gene ontology, KEGG: Kyoto encyclopedia of genes and genomes.
si-SREBF1 inhibits COAD cell migration and promotes autophagy
Cell migration and invasion are crucial for various physiological processes, such as cell development and immune defense, and are markers of the malignant progression of tumors. To further investigate the effect of silencing SREBF1 in COAD cells, HCT-116 and SW480 cells were transfected using si-SREBF1#2 and si-SREBF1#3, respectively, and found that silencing SREBF1 could significantly inhibit the horizontal and vertical COAD cell migration abilities through wound healing and Transwell assays [Figure 5a and b]. Autophagy plays an important role in maintaining intracellular environmental stability, coping with stress, and maintaining energy balance. Western blot results confirmed the inhibition effect of SREBF1 on the autophagy of COAD cells. Meanwhile, AO staining results similarly showed the presence of autophagosomes. These data indicate that inhibiting SREBF1 could induce autophagy of COAD cells. It has been demonstrated that this process increases the expression of autophagy markers, including Beclin-1 and LC3B, and also shows the presence of orange autophagosomes [Figure 5c and d, Supplementary Figure 1d and e], indicating that SREBF1 induces autophagy in COAD cells. IF staining also confirmed these findings [Figure 5e].
![Silencing Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) can inhibit the migration of colon adenocarcinoma cells and induce autophagy of these cells. (a and b) The effect of SREBF1 silencing on the migration ability of colon adenocarcinoma cells was measured by wound healing and Transwell assays. (c) Detection of the autophagy markers before and after silencing SREBF1 through western blot, and GAPDH was used as a loading control. (d) Acridine orange staining was used to observe the formation of autophagosomes. Magnification: ×200. (e) Immunofluorescence staining of LC3-II in constructed colon adenocarcinoma cells. [Blue: 4’,6-Diamidino-2’-phenylindole (DAPI); Green: Light chain (LC) 3-II], Magnification: ×400. The above experiments underwent replication 3 times (n = 3; ✶✶P < 0.01). GAPDH: Glyceralde-hyde-3-phosphate dehydrogenase.](/content/105/2024/21/1/img/Cytojournal-21-67-g005.png)
- Silencing Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) can inhibit the migration of colon adenocarcinoma cells and induce autophagy of these cells. (a and b) The effect of SREBF1 silencing on the migration ability of colon adenocarcinoma cells was measured by wound healing and Transwell assays. (c) Detection of the autophagy markers before and after silencing SREBF1 through western blot, and GAPDH was used as a loading control. (d) Acridine orange staining was used to observe the formation of autophagosomes. Magnification: ×200. (e) Immunofluorescence staining of LC3-II in constructed colon adenocarcinoma cells. [Blue: 4’,6-Diamidino-2’-phenylindole (DAPI); Green: Light chain (LC) 3-II], Magnification: ×400. The above experiments underwent replication 3 times (n = 3; ✶✶P < 0.01). GAPDH: Glyceralde-hyde-3-phosphate dehydrogenase.
Correlation between SREBF1 and immune infiltration level of COAD
The correlation between SREBF1 and immune infiltrating cells in the microenvironment of COAD was evaluated by the TIMER database, and the results indicate a statistically significant positive correlation between the presence of immune cells in COAD and variations in CNA of SREBF1 [Figure 6a]. Meanwhile, the TIMER database demonstrated SREBF1 expression levels with immune cells, B cells (partial cor = 0.074, P = 1.40e-01), CD8+T cells (partial cor = 0.05, P = 3.12e-01), CD4+T cells (partial cor = 0.127, P = 1.10e-02), neutrophil cells (partial cor = 0.177, P = 3.78e-04), and dendritic cells (partial cor = 0.264, P = 7.36e-08) in COAD cells [Figure 6b and c]. Furthermore, the Kaplan–Meier analysis demonstrated a significant association between survival outcomes and decreased infection levels in COAD patients with CD8+T cell (log-rank P = 0.023) and neutrophil (log-rank P = 0.044, Figure 6d) infiltration.
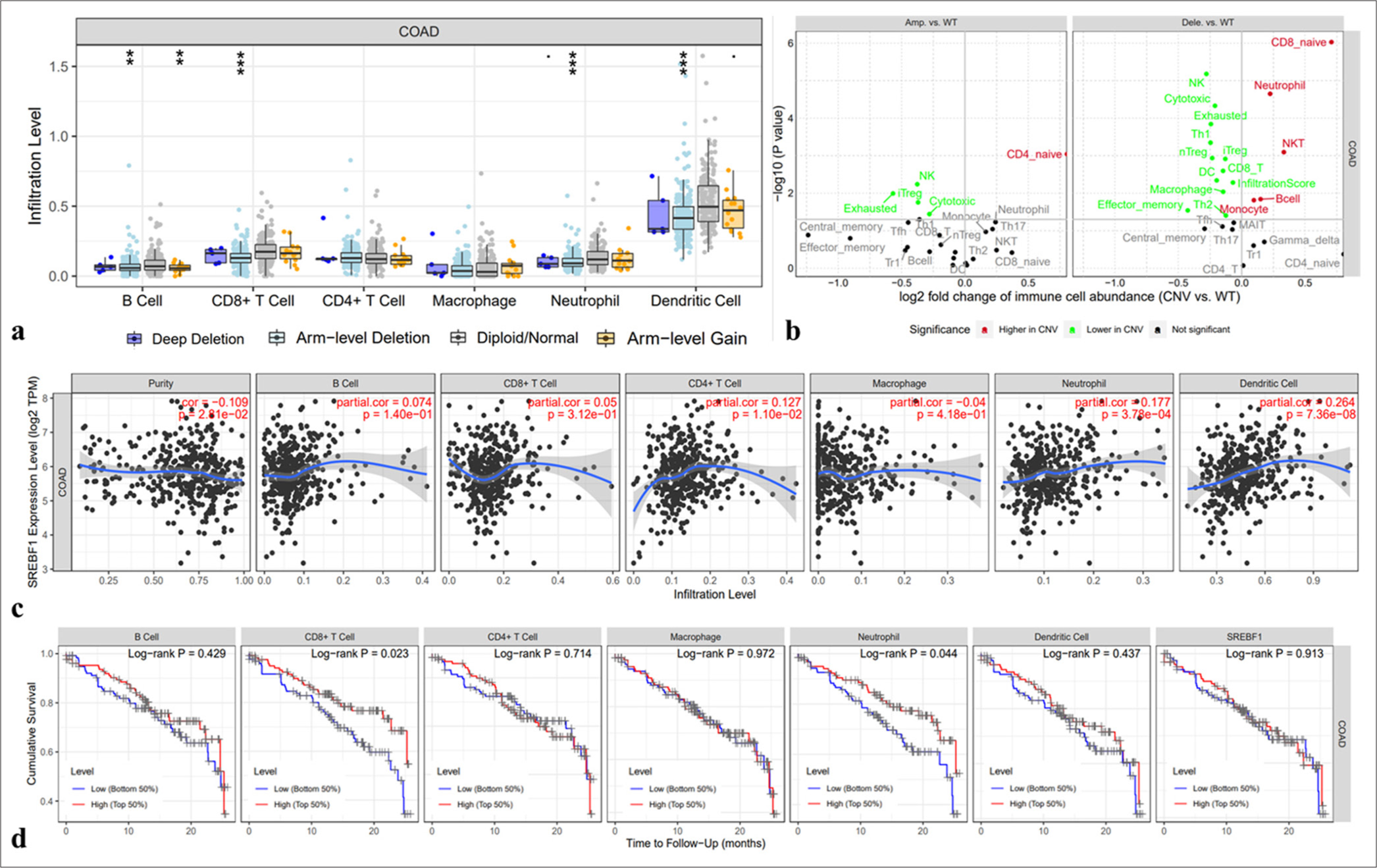
- The correlation between the expression of Sterol Regulatory Element Binding Transcription Factor 1 (SREBF1) and immune cells infiltration in colon adenocarcinoma. (a) The arm-level deletion probably inhibits the infiltration of immune cells. (b and c) Relationship between SREBF1 expression and infiltration of immune cells in colon adenocarcinoma from the TIMER database (A positive correlation exists between the expression of SREBF1 and infiltrating levels of B cell (r = 0.074, P = 1.40e-01), CD8+ T cells (r = 0.05, P = 3.12e-01), CD4+ T cell (r = 0.127, P = 1.10e-02), neutrophils (r = 0.177, P = 3.78e-04), and DCs (r =0.264, P = 7.36e-08) in colon adenocarcinoma). (d) Cumulative survival is related to CD8+ T-cell and neutrophil infiltration in colon adenocarcinoma (✶✶P < 0.01 and ✶✶✶P < 0.001).
DISCUSSION
SREBF1 is a gene responsible for encoding a protein that has a crucial function in the metabolism of cholesterol and lipids.[25] SREBF1 is a member of the Sterol Regulatory Element Binding Protein (SREBP) family, a transcription factor that plays a pivotal role in regulating the expression of numerous genes, particularly those involved in cholesterol and lipid synthesis.[26] SREBF1 primarily functions to control the production of cholesterol and lipids by attaching to the promoter regions of genes involved in their synthesis, thereby enhancing their transcription.[27] In this manner, SREBF1 helps maintain the balance of cholesterol and lipids in cells to meet their physiological needs. Many reports have demonstrated that SREBF1 is highly expressed in various cancers.[28] Li et al. demonstrated that the interaction between SREBF1 and key transcription factors plays a pivotal role in regulating cancer lipid metabolism and promoting tumor growth.[9] Database results also suggested that SREBF1 has significant upregulation in many cancer types, hence offering novel targets for clinical intervention in patients.
The relationship between genetic mutations and cancer is complex and multifaceted.[29] Genetic mutations can affect normal cell growth and apoptosis, increasing the risk of cancer. MSI, TMB, and neoantigens play important roles in cancer.[30,31] Liu et al. identified a significant correlation between the expression level of KIF18A and a number of key biological features, including immune cell infiltration, immune regulatory genes, immunological checkpoints, TMB, MSI, and mismatch repair (MMR), across a range of different malignancies.[32] Rodriquenz et al. discovered subpopulations of cancer that are positive for MSI and Epstein-Barr virus and identified their correlation with advanced immunotherapy. This finding offers valuable insights for the treatment of cancer patients.[33] MSI, TMB, and new antigens are widely used in personalized therapy and immunotherapy, providing better options for individualized treatment of patients.[34] In this study, through public database analysis, we found that SREBF1 has a high mutation frequency, with the most common mutation site being R586H/C. However, the clinical research value of this site mutation still needs to be studied. Especially in COAD, there is a robust link between the expression of SREBF1 and the presence of MSI, TMB, and neoantigens. Nevertheless, the precise connection between SREBF1 and COAD immune cell infiltration function remains uncertain, necessitating additional investigation.
Abnormal lipid signaling pathways may lead to cellular loss of control, thereby promoting the occurrence of cancer.[35] Our study demonstrates that SREBF1 exhibits high expression levels in multiple types of malignancies, with particular emphasis on COAD. Similarly, results from gene ontology (GO)-enriched and Kyoto Encyclopedia of Genes and Genomes (KEGG)-enriched databases showed that high expression of SREBF1 was significantly enriched in multiple biological functions, such as lipid metabolism-related processes in COAD. These findings are consistent with research by Zhang et al., which showed that SREBF1 increases the progression of esophageal squamous cell carcinoma through enhanced fatty acid synthesis.[36] Yang’s et al. research is consistent with our GO and KEGG analysis results, indicating that SREBP-1 may be affected by exposure to BPA during pregnancy and may be closely related to lipid metabolism disorders in later years.[37] SREBF1 is a pivotal transcription factor that predominantly controls lipid metabolism and has a significant impact on different tumor tissues.[38] Nevertheless, the specific function and mechanism of SREBF1 in COAD, especially its potential role through autophagy, is currently unclear. This study hypothesizes that SREBF1 may affect the development of COAD by regulating autophagy. To verify this hypothesis, we knocked down SREBF1 using siRNA transfection technology and detected its impact on autophagy levels. Through the utilization of western blot, specifically examining the LC3-II/I ratio, and IF staining of autophagosomes, we have discovered that the suppression of SREBF1 leads to a notable enhancement in the expression of autophagy associated markers, namely, LC3-II and Beclin-1. These results reveal that SREBF1 may affect the cellular behavior of colorectal cancer by inhibiting autophagy, and knocking down SREBF1 can relieve this inhibition and induce autophagy. Therefore, additional investigation is required to ascertain if SREBF1 controls the proliferation, differentiation, and cell death of COAD cells through lipid metabolism mechanisms.
The immune microenvironment is a key determinant of malignant progression and cancer treatment response,[39] it includes various immune cells, cytokines, chemicals, and other immune-related molecules that interact and affect tumor growth, spread and treatment response.[40-42] SREBF1 expression was correlated with immune cell presence in different tumors, especially CD8+ T-cells and neutrophils. Liu et al. found that regulatory T-cells inhibit CD8+ T-cell production of interferon γ, similar to our study, thereby enhancing the metabolic flexibility of tumor-promoting macrophages through a mechanism involving SREBP1.[43] Yang et al. also found that Ligustilide (Lig) significantly regulated lipid accumulation in alcohol exposed alpha mouse liver 12 cells through modulating peroxisome proliferator activated receptor alpha and SREBP1.[44] The abnormal expression of SREBP1 may affect the distribution and activity of immune cells in tumors.[45] However, there has not been extensive research on the specific association between SREBF1 and immune cell infiltration in COAD. This study showed a statistically significant positive correlation between the presence of immune cells in COAD and variations in CNA of SREBF1, but further experimental confirmation is needed. Moreover, it is worth mentioning that the expression of microsatellite instability MSI and SREBF1 may have an important relationship in COAD. MSI is a condition that arises from abnormalities in the DNA MMR machinery, resulting in genetic instability, which is common in COAD and is associated with specific clinical features and prognosis.[46] The results of the database suggest a possible correlation between the expression of SREBF1 in COAD and the status of MSI, indicating that MSI may affect the expression of SREBF1 through metabolic pathways. SREBF1 may act as a potential therapeutic target for MSI high status COAD, and the combined detection of MSI and SREBF1 expression may be helpful to predict the prognosis and treatment response more accurately. Therefore, understanding the relationship between MSI and SREBF1 in COAD has important clinical significance.
SUMMARY
SREBF1 is overexpressed in pan-cancer tissues. The overexpression of SREBF1 predicts the poor prognosis of the patients with COAD, and the variations in CNA of SREBF1 are positively correlated with the presence of immune cells in COAD. Meanwhile, silencing of SREBF1 could significantly inhibit the proliferation and migration of COAD cells, yet induce the autophagy of the cells. These results indicate that SREBF1 might be a novel approach to target therapy for COAD.
ACKNOWLEDGEMENTS
We are very grateful for the assistance provided by Yanbian University Central Laboratory.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
ABBREVIATIONS
AO – Acridine orange staining
CCK8 – Cell counting kit-8
CNA – copy number alteration
COAD – Colon adenocarcinoma
EdU – 5-ethynyl-2’-deoxyuridine
GAPDH – Glyceralde-hyde-3-phosphate dehydrogenase
IF – Immunofluorescence
IHC – Immunohistochemical staining
MSI – Microsatellite instability
PBS – Phosphate-buffered saline
PVDF – Polyvinylidene fluoride
si-RNA – Short interfering RNA
SREBF1 – Sterol Regulatory Element Binding Transcription Factor 1
TMB – Tumor mutation burden
TME – Tumor microenvironment
AUTHOR CONTRIBUTIONS
AHJ was involved in conception and design of the study, LYJ performed the experiments and data analysis; AHJ and ZHL performed drafting of the manuscript. All authors listed approved the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The research/study approved by the Institutional Review Board at Shanghai Outdo Biotech Company, number YB M-05-02, dated 2019.10.11. The authors certify that they have obtained all appropriate patient consent.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
EDITORIAL/PEER REVIEW
To ensure the integrity and highest quality of CytoJournal publications, the review process of this manuscript was conducted under a double-blind model (authors are blinded for reviewers and vice versa) through an automatic online system.
FUNDING
Not applicable.
References
- A bioinformatics approach to identify a disulfidptosis-related gene signature for prognostic implication in colon adenocarcinoma. Sci Rep. 2023;13:12403.
- [CrossRef] [PubMed] [Google Scholar]
- A novel enterocyte-related 4-gene signature for predicting prognosis in colon adenocarcinoma. Front Immunol. 2022;13:1052182.
- [CrossRef] [PubMed] [Google Scholar]
- Trends in targeted delivery of nanomaterials in colon cancer diagnosis and treatment. Med Res Rev. 2022;42:227-58.
- [CrossRef] [PubMed] [Google Scholar]
- Hidden colon adenocarcinoma diagnosed from mouth metastasis: Case report and literature review. World J Surg Oncol. 2023;21:88.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical updates for colon cancer care in 2022. Clin Colorectal Cancer. 2022;21:198-203.
- [CrossRef] [PubMed] [Google Scholar]
- Integrative deep learning analysis improves colon adenocarcinoma patient stratification at risk for mortality. EBioMedicine. 2023;94:104726.
- [CrossRef] [PubMed] [Google Scholar]
- Construction of a novel choline metabolism-related signature to predict prognosis, immune landscape, and chemotherapy response in colon adenocarcinoma. Front Immunol. 2022;13:1038927.
- [CrossRef] [PubMed] [Google Scholar]
- SREBF1/SREBP-1 concurrently regulates lipid synthesis and lipophagy to maintain lipid homeostasis and tumor growth. Autophagy. 2024;20:1183-5.
- [CrossRef] [PubMed] [Google Scholar]
- Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat Commun. 2021;12:4362.
- [CrossRef] [PubMed] [Google Scholar]
- Sterol regulatory element binding transcription factor 1 promotes proliferation and migration in head and neck squamous cell carcinoma. PeerJ. 2023;11:e15203.
- [CrossRef] [PubMed] [Google Scholar]
- The NQO1/p53/SREBP1 axis promotes hepatocellular carcinoma progression and metastasis by regulating Snail stability. Oncogene. 2022;41:5107-20.
- [CrossRef] [PubMed] [Google Scholar]
- Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023;41:404-20.
- [CrossRef] [PubMed] [Google Scholar]
- Aspects of the tumor microenvironment involved in immune resistance and drug resistance. Front Immunol. 2021;12:656364.
- [CrossRef] [PubMed] [Google Scholar]
- Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer. 2022;8:28-42.
- [CrossRef] [PubMed] [Google Scholar]
- TGF-β signaling in the tumor metabolic microenvironment and targeted therapies. J Hematol Oncol. 2022;15:135.
- [CrossRef] [PubMed] [Google Scholar]
- The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93-9.
- [CrossRef] [PubMed] [Google Scholar]
- TIMER v2.0. U.S. Dana farber cancer institute. Available from: https://cistrome.shinyapps.io/timer [Last accessed on 2024 Mar 20]
- [Google Scholar]
- UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649-58.
- [CrossRef] [PubMed] [Google Scholar]
- Gene set cancer analysis (GSCA) China: Hu Lab, School of Medicine, WUST; Available from: http://bioinfo.life.hust.edu.cn/GSCA [Last accessed on 2024 Mar 20]
- [Google Scholar]
- Gene expression profiling interactive analysis (GEPIA) China: Zhang's Lab; from Available from: https://gepia.cancer-pku.cn [Last accessed on 2024 Mar 20]
- [Google Scholar]
- The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4.
- [CrossRef] [PubMed] [Google Scholar]
- SangerBox. China: SangerBox R and D Department; Available from: https://sangerbox.com/home.html [Last accessed on 2024 Mar 20]
- [Google Scholar]
- The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638-46.
- [CrossRef] [PubMed] [Google Scholar]
- World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191-4.
- [CrossRef] [PubMed] [Google Scholar]
- CD36 promotes de novo lipogenesis in hepatocytes through INSIG2-dependent SREBP1 processing. Mol Metab. 2022;57:101428.
- [CrossRef] [PubMed] [Google Scholar]
- Regulatory roles of SREBF1 and SREBF2 in lipid metabolism and deposition in two Chinese representative fat-tailed sheep breeds. Animals (Basel). 2020;10:1317.
- [CrossRef] [PubMed] [Google Scholar]
- Macrophage SREBP1 regulates skeletal muscle regeneration. Front Immunol. 2024;14:1251784.
- [CrossRef] [PubMed] [Google Scholar]
- Regulation and targeting of SREBP-1 in hepatocellular carcinoma. Cancer Metastasis Rev. 2024;43:673-708.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic mutations affecting mitochondrial function in cancer drug resistance. Genes Genomics. 2023;45:261-70.
- [CrossRef] [PubMed] [Google Scholar]
- A multi-omics analysis reveals CLSPN is associated with prognosis, immune microenvironment and drug resistance in cancers. Biol Proced Online. 2023;25:16.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125-34.
- [CrossRef] [PubMed] [Google Scholar]
- Comprehensive pan-cancer analysis of KIF18A as a marker for prognosis and immunity. Biomolecules. 2023;13:326.
- [CrossRef] [PubMed] [Google Scholar]
- MSI and EBV positive gastric cancer's subgroups and their link with novel immunotherapy. J Clin Med. 2020;9:1427.
- [CrossRef] [PubMed] [Google Scholar]
- ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann Oncol. 2019;30:1232-43.
- [CrossRef] [PubMed] [Google Scholar]
- Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr Opin Cell Biol. 2020;63:57-67.
- [CrossRef] [PubMed] [Google Scholar]
- PRP19 enhances esophageal squamous cell carcinoma progression by reprogramming SREBF1-dependent fatty acid metabolism. Cancer Res. 2023;83:521-37.
- [CrossRef] [PubMed] [Google Scholar]
- Gestational bisphenol A exposure impairs hepatic lipid metabolism by altering mTOR/CRTC2/SREBP1 in male rat offspring. Hum Exp Toxicol. 2022;41:9603271221129852.
- [CrossRef] [PubMed] [Google Scholar]
- C9orf72 controls hepatic lipid metabolism by regulating SREBP1 transport. Cell Death Differ. 2024;31:1070-84.
- [CrossRef] [PubMed] [Google Scholar]
- Unlocking the potential of the tumor microenvironment for cancer therapy. Pathol Res Pract. 2023;251:154846.
- [CrossRef] [PubMed] [Google Scholar]
- Immunotherapy: Reshape the tumor immune microenvironment. Front Immunol. 2022;13:844142.
- [CrossRef] [PubMed] [Google Scholar]
- Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126-33.
- [CrossRef] [PubMed] [Google Scholar]
- Unexpected guests in the tumor microenvironment: Microbiome in cancer. Protein Cell. 2021;12:426-35.
- [CrossRef] [PubMed] [Google Scholar]
- Treg cells promote the SREBP1-dependent metabolic fitness of tumor-promoting macrophages via repression of CD8+ T cell-derived interferon-γ. Immunity. 2019;51:381-97.e6.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting RXFP1 by ligustilide: A novel therapeutic approach for alcoholic hepatic steatosis. Int Immunopharmacol. 2024;127:111460.
- [CrossRef] [PubMed] [Google Scholar]
- Impaired lipid biosynthesis hinders anti-tumor efficacy of intratumoral iNKT cells. Nat Commun. 2020;11:438.
- [CrossRef] [PubMed] [Google Scholar]
- Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics. 2020;10:11080-91.
- [CrossRef] [PubMed] [Google Scholar]








