Translate this page into:
Protein extraction from methanol fixed paraffin embedded tissue blocks: A new possibility using cell blocks
*Corresponding author
-
Received: ,
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background:
Methanol fixed and paraffin embedded (MFPE) cellblocks are an essential cytology preparation. However, MFPE cellblocks often contain limited material and their relatively small size has caused them to be overlooked in biomarker discovery. Advances in the field of molecular biotechnology have made it possible to extract proteins from formalin fixed and paraffin embedded (FFPE) tissue blocks. In contrast, there are no established methods for extracting proteins from MFPE cellblocks. We investigated commonly available CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) buffer, as well as two commercially available Qiagen® kits and compared their effectiveness on MFPE tissue for protein yields.
Materials and Methods:
MFPE blocks were made by Cellient™ automated system using human tissue specimens from normal and malignant specimens collected in ThinPrep™ Vials. Protein was extracted from Cellient-methanol fixed and paraffin embedded blocks with CHAPS buffer method as well as FFPE and Mammalian Qiagen® kits.
Results:
Comparison of protein yields demonstrated the effectiveness of various protein extraction methods on MFPE cellblocks.
Conclusion:
In the current era of minimally invasive techniques to obtain minimal amount of tissue for diagnostic and prognostic purposes, the use of commercial and lab made buffer on low weight MFPE scrapings obtained by Cellient® processor opens new possibilities for protein biomarker research.
Keywords
Cellblocks
Cellient
CHAPS buffer
extraction
formalin fixed and paraffin embedded
methanol fixation
methanol fixed and paraffin embedded
protein
Qiagen
INTRODUCTION
The traditional cellblock is an essential cytology preparation, which offers benefits over cytologic smears by preservation of cell architecture and the performance of immunohistochemical studies. In the current era of minimally invasive techniques for obtaining tissue samples for diagnostic and prognostic markers, cellblocks containing “limited material” specimens are routinely used to provide the valuable information about pre-disease and disease processes.
The methanol fixed and paraffin embedded (MFPE) cellblocks are prepared manually with minimal automation with their quality highly dependent upon the experience of the cytopreparation personnel. Currently, Cellient® automated cell block system is widely used for MFPE cellblock preparation to ensure consistent quality preparation by minimizing the operator dependency. Despite advances in technology, the relatively small size of the cytology scrapings MFPE cell blocks in comparison to the formalin fixed and paraffin embedded (FFPE) counterparts have caused them to be often overlooked in biomarker discovery.
Recently, in the field of proteomics, there is an emphasis on utilizing limited tissue samples such as core biopsies and cellblocks for the evaluation of molecular biomarkers. These have the potential of being eventually converted into novel diagnostic marker assays that will aid in improving the efficacy of clinical intervention, as well as the validation of leads and targets. Proteomic platforms have developed over the past few years due to advances in scientific knowledge and technology with the next technologic leap being the application of proteomic technologies to the bedside.[123] At present, there is a lack of established methods or resources for extracting proteins from MFPE cell blocks.
In the current study, we investigated the protein yield from MFPE cellblocks employing the Cellient™ automated cell block system. The Cellient™-methanol fixed and paraffin embedded (C-MFPE) blocks were prepared from four different tissue types. Protein was extracted from the C-MFPE blocks containing tissue and scrapings using commercial and lab-made CHAPS buffer; the concentration was estimated and protein yield (ugs) per milligram of tissues and scrapings were evaluated.
MATERIALS AND METHODS
Human tissue samples
The study was conducted in accordance with the policies stated by the University of Pennsylvania Institutional Review Board. Tissue samples were provided by the Cooperative Human Tissue Network (CHTN), which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects.
Normal (Stomach, Colon) (n = 8/tissue group) and malignant (Colon, Kidney) (n = 3-4/tissue group) tissue specimens were collected by CHTN. The malignant specimens included renal cell carcinomas and colorectal carcinomas.
Tissue processing and workflow
Both scrapings and small aliquots of the tissues from each specimen were obtained to quantify protein extraction per unit of material. The tissue scrapings were obtained and suspended into the ThinPrep® Non-Gyn vial. Following the collection of scrapings, aliquots of the tissue were obtained for (1) Snap-freezing, (2) FFPE, and (3) ThinPrep® Non-Gyn vial processing [Figure 1]. One of the aliquots were snap-frozen in the vapor phase of the liquid nitrogen while another was placed in the FFPE process that averaged tissue fixation in 10% phosphate buffered formalin - 2 h, followed-by tissue processing - 10 h and paraffin embedding - 20 min. The scrapings and the tissue aliquots collected into ThinPrep® vial containing Preservcyte® were fixed overnight and processed into a Cellient® block per manufacturer directions. These processed blocks containing the scrapings and tissues were termed as C-MFPE tissue scraping blocks and as C-MFPE tissue blocks respectively. The blocks were stored at room temperature until processing.
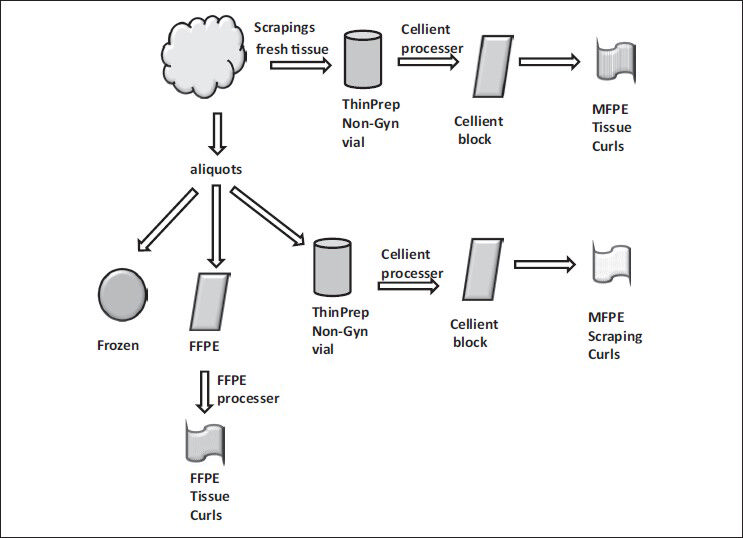
- Processing of freshly obtained tissue
Protein extraction from C-MFPE samples
C-MFPE samples (both scrapings and tissues) processed using automated Cellient® cell block system was sectioned into 5-7 um tissue curls. These were deparaffinized in xylene, washed in ethanol followed-by rehydration in phosphate buffered saline (PBS) and centrifuged at 14,000 g for 30 s. The resulting pellet was divided into three aliquots [Figure 2]. The aliquots were weighed and suspended in (1) Qproteome FFPE Tissue Kit (Qiagen Corp, CA, USA) or (2) Qproteome Mammalian Tissue kit (Qiagen Corp, CA, USA) or (3) Optimized CHAPS lysis buffer (30 mM Tris-Cl pH 7.5, 150 mM NaCl, 1% CHAPS, 0.03 mM dithiothreitol, protease inhibitor cocktail). The suspension was taken through 5-6 cycles of freeze-thaw to ensure maximum tissue lysis. The freezing was performed on dry ice for 5 min followed-by 10 min of thawing at 37°C. The freeze-thawed suspension was homogenized using TissueLyser LT (Qiagen Corp, CA, USA). To ensure maximum protein extraction, the homogenized tissues were first heated at 90°C for 20 min followed-by heating (for 2 h for tissues in FFPE buffer and 1 h for tissues in mammalian tissue buffer, as well as CHAPS buffer) at 80°C. The resultant lysate was centrifuged at full speed for 10 min and the supernatant was taken out for bicinchoninic acid (BCA) assay.

- Workflow for extraction of protein using various buffers from Cellient- methanol fixed and paraffin embedded tissue block curls
Protein extraction from FFPE tissue
The 5-7 um tissue curls obtained from the FFPE blocks were deparaffinized followed-by hydration as above, centrifuged at 14,000 g for 30 s and weighed. Approximately 50-100 mg of sectioned sample was used for protein extraction using the kit protocol of Qproteome FFPE Tissue (Qiagen Corp, CA, USA). Protein was extracted in 100 ul of extraction buffer (EXB) and the concentration was determined using BCA assay.
Protein extraction from frozen samples
Briefly, approximately 30 mg of frozen tissue stored at −80°C was weighed, further cooled in liquid nitrogen and freeze-fractured in Covaris TT1® tissue tubes using the Covaris CryoPrep™ pulverization system at an impact setting of 5. The pulverized tissues were transferred to TissueLyser LT (Qiagen Corp, CA, USA) and homogenized for 5 min. The protein extraction was accomplished by using the kit protocol of Qproteome mammalian kit (Qiagen Corp, CA, USA). Protein was extracted in 1000 ul of frozen EXB and the concentration was determined using BCA assay.
Analysis
Concentration of the total extracted protein was obtained using BCA assay. To allow for comparison between methods, total extracted protein was then normalized to the weight of tissue to obtain “microgram of protein per milligram of tissue.” The weights used in this calculation are weights obtained once the curls were deparaffinized in xylene and rehydrated with PBS for C-MFPE and FFPE tissue curls while for the frozen tissue, it was the weight prior to cooling for freeze fracture pulverization. The resulting protein unit of “microgram of protein per milligram of tissue” for various methods was analyzed for statistical significance using Students t-test.
RESULTS
The results are summarized in Tables 1–3.

The processed blocks containing the scrapings and tissues were termed as C-MFPE scraping blocks and as C-MFPE tissue blocks respectively. A representative image of an H and E stained C-MFPE scraping block section and C-MFPE tissue block shows cellular spread across section [Figure 3].
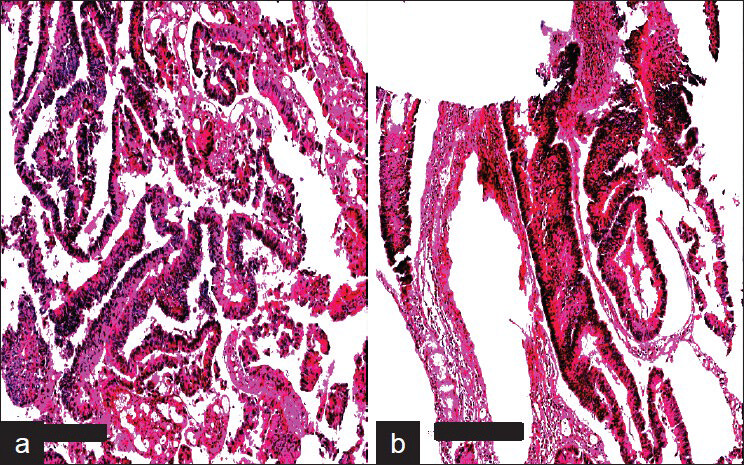
- Aperio Images of adenocarcinoma colon stained with H&E. a) MFPE Cellient block prepared from tissue and b) MFPE block prepared from scrapings collected in ThinPrep vial. Scale bar represents 300um.
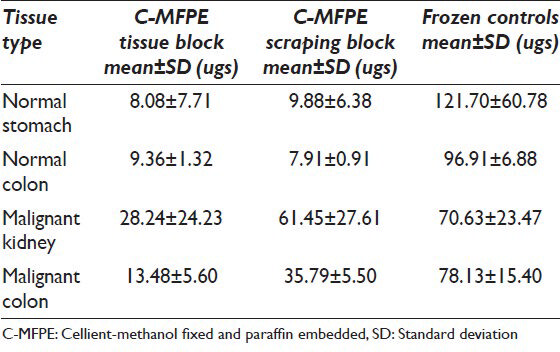
Tissue and scraping from C-MFPE blocks were processed using one of the following methods: Qiagen's FFPE method, Qiagen's mammalian method and CHAPS lysis buffer method. Protein extraction using FFPE method showed that the protein yields per milligram of tissue were highest for C-MFPE scraping blocks, followed by C-MFPE tissue blocks with the lowest for control FFPE tissue blocks. This result is interesting since, FFPE kit formulated for FFPE blocks was expected to have a higher yield than C-MFPE blocks [Table 1]. In contrast, use of Qiagen's mammalian buffer on frozen controls yielded high protein yield as expected. In comparison with their frozen tissue counterparts, there were approximately three to ten-fold decreases in the yields from C-MFPE blocks [Table 2]. An exception was the yield from malignant kidney C-MFPE scraping block, which was comparable to the yield from frozen tissue. CHAPS lysis buffer commonly used for protein extraction was evaluated directly on the C-MFPE tissue/scraping blocks [Table 3].
Comparable yields were obtained with all three methods across C-MFPE for colorectal carcinoma, normal colon and normal stomach tissue blocks. However, for renal cell carcinoma C-MFPE tissue block, similar yields were obtained with FFPE and frozen methods while a three-fold decrease in yield was observed with CHAPS method [Figure 4]. Higher protein yields were observed across C-MFPE scraping blocks in comparison to C-MFPE tissue blocks. Similarly, for normal stomach, FFPE method gave the highest yield while for malignant kidney highest yields were obtained using both frozen and FFPE methods [Figure 5].

- Comparison of protein yields obtained from C-MFPE tissue blocks
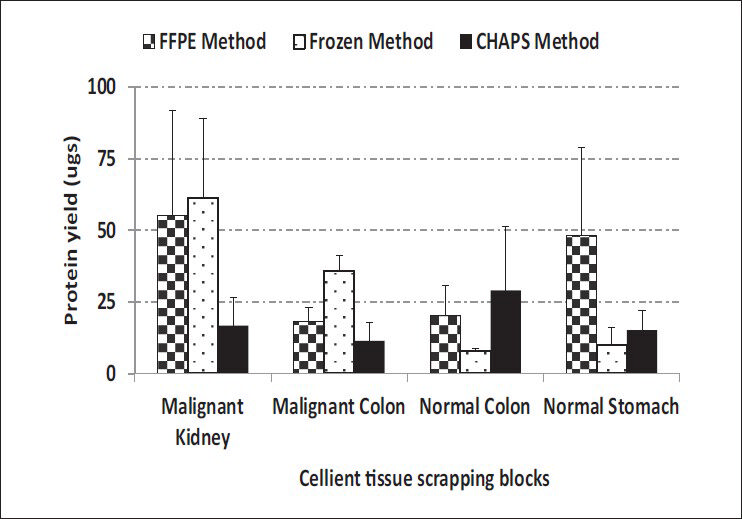
- Comparison of protein yields obtained from C-MFPE scraping blocks
Graphical representation of protein yield (ug) per unit starting material for the samples used for each method have been plotted as a scatter plot in Figure 6 (C-MFPE scraping blocks) and Figure 7 (C-MFPE tissue blocks).
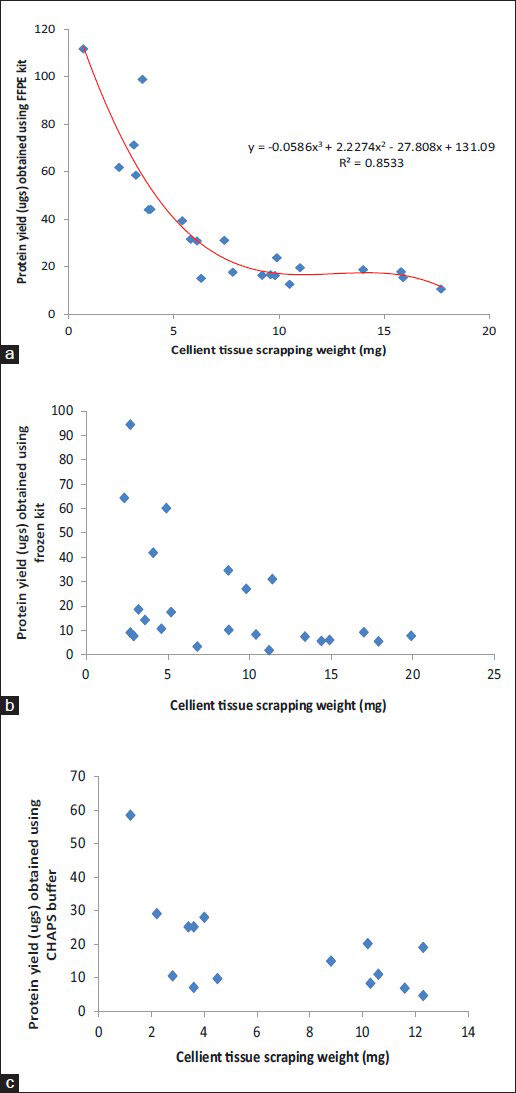
- Comparison analysis of protein yields obtained using (a) formalin fixed and paraffin embedded method, (b) mammalian kit method kit and (c) CHAPS buffer from Cellient-methanol fixed and paraffin embedded tissue scraping blocks

- Comparison analysis of protein yields obtained using (a) formalin fixed and paraffin embedded method, (b) mammalian kit method kit and (c) CHAPS buffer from Cellient-methanol fixed and paraffin embedded tissue blocks
DISCUSSION
MFPE cell blocks are an essential cytology preparation that provides architectural and cytologic details similar to those seen in small tissue biopsy specimens.[4] The cellblock's tissue sections allow for multiple immunohistochemical staining for a battery of markers. Thus, they are often preferred over other cytologic preparations such as cytospins and smears for immunohistochemical analysis.[567] However, there are no established methods or resources for extracting proteins from MFPE cellblocks prepared from the cytopathology specimens containing very limited amount of diagnostic tissue.
In this study, we investigated a new method for the extraction of proteins from MFPE cellblocks prepared on the Cellient™ automated cell block system. This was accomplished by using commonly available CHAPS buffer and two commercially available kits on four different tissue types. The end result of our investigations is a ‘total protein extract’, which was quantified for comparison using BCA assay. The protein kits that were investigated were chosen on their established ability to extract protein from the correlated preparation type (i.e. mammalian buffer on frozen tissues and FFPE buffer on FFPE tissues). Thus, usage of these kits as well as the use of CHAPS lysis buffer for protein extraction on MFPE processed tissues is novel.
The goal of this current investigation is to show that protein extraction is feasible from MFPE embedded tissues. In this study, we focused on ‘total protein extract’ from the tissues and will investigate the quality and purity of particular proteins of interest in a separate study. To this effect, we examined a wide range of starting material needed to quantify protein extraction per unit of starting material by using both scrapings and small chunks of tissues from each tissue. One of the challenges we faced was the homogenization of deparaffinized and rehydrated curls in the lysis buffer from MFPE tissue and scraping blocks. The lysis buffers used were 100 ul of EXB for FFPE protocol, 1000 ul of frozen buffer for frozen protocol, 250 ul of CHAPS lysis buffer for scrapings and 500 ul CHAPS lysis buffer for tissue curls. Once the tissues were in their respective buffers, they were taken through 5-6 cycles of freeze-thaw to ensure maximum tissue lysis. The freezing was performed on dry ice for 5 min followed-by 10 min of thawing at 37°C. This step was then followed-by routine homogenization and extraction.
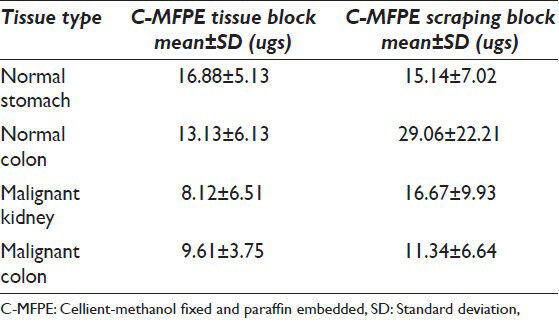
Comparison of protein yields for C-MFPE tissue blocks using frozen, FFPE kits and CHAPS buffers show a comparable protein yields, however, protein yield analysis of scrapings showed highest yields. The increase in yields per unit of tissue in scraping blocks might be attributed to scrapings having more surface area than the tissue; however, the yields for different methods were tissue specific. For instance, in the case of colorectal carcinoma, highest protein yield was obtained with frozen method while in the case of normal colon, CHAPS methods gave the highest yield.
An important finding our study showed is the minimum requirement of tissue weight that can be used to extract the proteins. For FFPE method, the lowest weight used was 0.7 mg scraping from normal stomach corresponding to 111 ugs of protein/mg of tissue. Meanwhile, for mammalian buffer, 2.7 mg malignant kidney scraping was the lowest weight used corresponding to a protein yield of 94 ugs of protein/mg of tissue. The lowest weight of scraping used with CHAPS lysis buffer was 1.2 mg of normal colon corresponding to a protein yield of 58 ugs of protein/mg of tissue. Analysis of weight and protein yield for C-MFPE tissue and scraping block for FFPE, mammalian and CHAPS buffers are represented in Figures 6 and 7. In both scraping, as well as tissue block, a polynomial relationship exists for FFPE method while R2 value of trend observed for mammalian and CHAPS buffer was below 0.3.
CONCLUSION (S) AND SUMMARY
Our investigation assesses the feasibility employing small tissue samples to obtain total protein, which can be employed for biomarker discovery. We demonstrate the efficacy of various protein extraction methods on C-MFPE cellblocks. Further assessment of the acquired proteins through Western Blot and other methods are underway for a separate investigation. In addition, the MFPE cellblocks can be stored at room temperature over long periods and can function as a veritable resource whose potential can be unlocked with the extraction of protein molecular derivatives. Thus, the use of commercial and lab-made buffer on low-weight MFPE scrapings obtained by Cellient® processor opens new possibilities for protein assay studies in the current era of minimally invasive techniques to obtain minimal amount of tissue for diagnostic and prognostic purposes.
COMPETING INTERESTS STATEMENT BY ALL AUTHORS
The author (s) declare that they have no competing interests.
AUTHORSHIP STATEMENT BY ALL AUTHORS
All authors of this article declare that we qualify for authorship as defined by ICMJE. Each author has participated sufficiently in the work and takes public responsibility for appropriate portions of the content of this article.
ZB conceived of the study, participated in its design and coordination and in the revision of the manuscript. VL participated in its design and coordination and helped to draft the manuscript. AT procured tissue and processed cellblocks carried out protein extraction experiments and helped to draft the manuscript. MP procured tissue and processed cellblocks, carried out protein extraction experiments and helped to draft the manuscript. DM participated in its design, coordination and helped to draft the manuscript. TK participated in its design, coordination, helped in carrying out protein extraction experiments, analyzed as well as interpreted the data and drafted the manuscript.
All authors read and approved the final manuscript. Each author acknowledges that this final version was read and approved.
ETHICS STATEMENT BY ALL AUTHORS
This study was conducted with approval from University of Pennsylvania Institutional Review Board (IRB). Authors take responsibility to maintain relevant documentation in this respect.
EDITORIAL/PEER-REVIEW STATEMENT
To ensure the integrity and highest quality of Cyto Journal publications, the review process of this manuscript was conducted under a double-blind mode.(authors are blinded for reviewers and vice versa) through automatic online system.
ACKNOWLEDGMENTS
We would like to acknowledge Ms. Gina Piermatteo for C-MFPE and FFPE curls. We would also like to acknowledge NCI for grant #CA-044974 provided to the Eastern Division of the cooperative human tissue network.
Available FREE in open access from: http://www.cytojournal.com/text.asp?2013/10/1/23/122299
REFERENCES
- Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2004;3:984-97.
- [Google Scholar]
- A modified cell block technique for fine needle aspiration cytology. Acta Cytol. 1995;39:93-9.
- [Google Scholar]
- Effectiveness of the cell block technique in diagnostic cytopathology. J Cytol. 2012;29:177-82.
- [Google Scholar]








