Translate this page into:
Abstracts for the 59th Annual Scientific Meeting (November 2011) by American Society of Cytopathology (ASC) at Baltimore, MD, USA
-
Received: ,
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Abstract
These are peer-reviewed poster-platform submissions finalized by the Scientific Program Committee. A total of 153 abstracts (14 Platforms [PP1 through PP14] & 139 Posters [1 through 139]) were selected from 161 submissions to be considered for presentation during November 4 – 8, 2011, at the Hilton Baltimore Hotel, to pathologists, cytopathologists, cytotechnologists, residents, fellows, students, and other members of cytopathology-related medical and scientific fields.
Keywords
Abstracts
American society of cytopathology
ASC
cytopathology
cytology
PP 1: Can high risk human papillomavirus positivity in a negative for intraepithelial lesion or malignancy in women 30 years of age and older be used to select cases for quality control review?
Sarah Wehmhoefer, CT(ASCP), Michael Beckmann, DO
Department of Laboratory Medicine and Pathology, Memorial Medical Center, Springfield, Illinois
Introduction: This study is aimed to evaluate if women aged 30 years or older, who are Pap and high-risk human papillomavirus (HR HPV) co-tested and found to be negative for intraepithelial lesion or malignancy (NILM) and positive for HR HPV, would benefit from a retrospective Pap re-screen.
Materials and Methods: Following institutional review board approval, a search of the cytopathology laboratory database from January to December, 2010, yielded 55975 Pap tests. One thousand seven hundred and six (3%) patients aged 30 years or older (range 30 – 61) were found to have been co-tested with a SurePath® Pap and HR HPV. High risk HPV testing was performed using Qiagen (Digene) Hybrid Capture 2 (HC2). Of those 1706, 72 (4%) patients who had an NILM Pap and were positive for HR HPV were re-screened by a Quality Control (QC) cytotechnologist (SMW) with the knowledge of the HR HPV status. Those cases determined to be abnormal upon re-screening were evaluated by a cytopathologist (MJB) who determined the final re-screen interpretation.
Results: Of the 72 re-screened cases, 12 cases (17%) were found to be atypical squamous cells (ASC) or higher on the final re-interpretation. Six cases were found to be atypical squamous cells of undetermined significance (ASC-US), two Atypical Squamous Cells, cannot exclude high grade squamous intraepithelial lesions (ASC-H), two low-grade squamous intraepithelial lesions (LSIL), two low-grade squamous intraepithelial lesions, Cannot exclude high-grade squamous intraepithelial lesions (LSIL-H), and 0 high-grade squamous intraepithelial lesions (HSIL) or atypical glandular cells (AGC). Given the cytopathology laboratory use of the focal point primary screening instrument, 40% of all Pap tests advanced to a QC queue. Of those QC Pap tests, 1.5% were upgraded to ASC or higher upon re-screening in 2010.
Conclusions: The focused re-screening of NILM Pap tests with positive HR HPV of women 30 years and older showed a higher detection rate of ASC or higher compared to routine QC screening. The current American society for colposcopy and cervical pathology (ASCCP) consensus guidelines recommend repeating both the Pap test and HR HPV test in one year if a woman aged 30 years or older has an NILM Pap and positive HR HPV test. If the follow-up Pap test is again NILM and HR HPV positive, colposcopy is suggested. This algorithm may change in the future, but focused re-screening in this patient population may provide an additional QC measure in the interim.
PP 2: The tahoe study: Bias in interpretation of Pap tests when HPV status is known
Ann Moriarty, MD1, Ritu Nayar, MD2, Andrew Renshaw, MD3, Nicole Thomas, MPH, CT(ASCP)4, Rhona Souers, MS5
1Pathology, AmeriPath, Indianapolis, Indiana; 2Pathology, McGaw Medical Center of Northwestern University, Chicago, Illinois; 3Pathology, Baptist Health, Miami, Florida; 4Surveys, College of American Pathologists, Northfield, Illinois; 5Biostatistics, College of American Pathologists, Northfield, Illinois
Introduction: The performance characteristics of cervical cytology as a primary screening test are well known. However, little data is available on the performance of Pap tests if the human papillomavirus (HPV) status is known prior to screening. The potential bias of the HPV status may affect the Pap test performance if HPV molecular testing is used as an initial screening test for cervical cancer followed by Pap testing.
Materials and Methods: Forty de-identified liquid-based Pap tests were selected from a quality assurance (QA) program that reviewed negative Pap tests after a positive HPV test. They were divided into two groups of 20 slides and circulated among two randomly assigned groups of 22 members, from the College of American Pathologist (CAP) Cytopathology Committee (CYP), who categorized them according to the Bethesda System (TBS). The slides were re-labeled and each batch was circulated to the opposite group, who also categorized them using TBS, after being told that all cases were HPV positive. Each response was evaluated against the general diagnosis category of negative for intraepithelial lesion (NILM) and epithelial cell abnormality (ECA), as well as, the descriptive diagnostic categories of atypical squamous cells of undetermined significance (ASC-US), low-grade squamous intraepithelial lesion (LSIL) or high-grade squamous intraepithelial lesion (HSIL). Differences in the responses between groups were analyzed statistically by the chi-square and Cochran-Mantel-Haenszel tests at the 0.05 significance level.
Results: The unbiased group were more likely to identify the slides as negative for intraepithelial lesion or malignancy (NILM) and less likely to identify an epithelial cell abnormality (ECA) than the biased group (P < 0.001), as depicted in Table 1. There was an increase in each descriptive diagnostic category of epithelial cell abnormalities in the biased responses as compared to the unbiased responses (P = 0.002), as seen in Table 2.
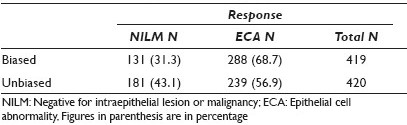
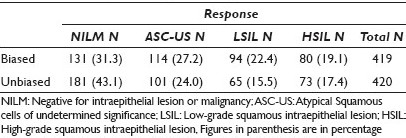
Conclusions: Knowledge of HPV status creates a bias in the interpretation of Pap tests. When the HPV status is known, observers are more likely to report the Pap test as abnormal and with greater frequency in all categories of epithelial cell abnormality (ASC-US, LSIL, and HSIL). If molecular testing is used as a primary screening test and Pap tests are used as secondary or reflex tests, it is more likely that the Pap test will be interpreted as abnormal, as compared to when a Pap test is used as a primary screening test.
PP 3: Risk profiling using HPV genotyping in women with mildly abnormal Pap results
Ming Guo, MD1, Jianping Wang, CT(ASCP)1, Marilyn Dawlett, CT(ASCP)1, Shobha Patel, CT(ASCP)1, Ping Liu2, Yun Gong, MD1, Therese Bevers, MD3, Nour Sneige, MD1
1Pathology, UT MD Anderson Cancer Center, Houston, Texas; 2Biostatistics, UT MD Anderson Cancer Center, Houston, Texas; 3Cancer Prevention Center, UT MD Anderson Cancer Center, Houston, Texas
Introduction: HPV DNA testing is the standard of care in triage of women with atypical squamous cells of undetermined significance (ASC-US) Pap results. Genotyping for HPV types16 / 18 was recommended by the American Society for Colposcopy and Cervical Pathology (ASCCP) for women with Pap- / HPV+ results. It is unknown whether adding a genotyping test for HPV 16 / 18 would improve patient management by risk profiling in women with mildly abnormal Pap results, that is, ASCUS, or low-grade squamous intraepithelial lesion (LSIL). To answer this question, we performed a study to evaluate the predictive value of HPV genotyping for cervical intraepithelial lesion / vaginal intraepithelial neoplasm (CIN / VAIN) 2+ in women with mildly abnormal Pap results and HPV DNA+ testing results.
Materials and Methods: We collected SurePath® Pap specimens from 351 women at the Department of Pathology, MD Anderson Cancer Center from 2005 to 2008 that met the criteria of this study. From these, 240 specimens had ASCUS Pap results and 111 specimens had LSIL Pap results, all with Hybrid Capture (HC2) HPV testing results. HPV genotyping was performed on these using the EasyChip HPV assay, which can detect 39 HPV genotypes. The Pap specimens with HPV genotypes 16 / 18, high-risk non16 / 18 (HR-HPV), and low-risk HPV types were compared with the follow-up biopsy results. Follow-up duration ranged from 1 to 36 months with a mean of 21 months.
Results: Of the 351 women, 36 (10.3%) had HC2- results. Genotyping showed low-risk HPV in 2 cases (5.6%, 2 / 36). In 315 women with HC2+ results, 18 (5.7%) tested negative using the polymerase chain reaction (PCR) assay. The distribution of HPV genotypes and a comparison with the results of follow-up biopsies appear in Table 1. Women with ASCUS or LSIL Pap results and HPV16 / 18 had significantly higher rates of CIN / VAIN2 / 3 on follow-up biopsies than did women with low-risk HPV types (P = 0.01-0.02). Higher odds ratios (OR) for CIN / VAIN2+ was found in women who had HPV16 / 18 genotyping and ASCUS (13.9, 95% CI, 1.7-113.4) or LSIL (5.7, 95% CI, 1.4-24.4) Pap results compared with women who had low risk-HPV genotypes.
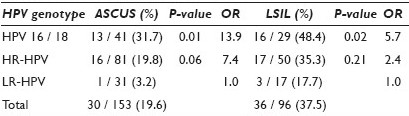
Conclusions: HPV genotyping may be useful for risk profiling in women with mildly abnormal Pap results, especially ASC-US. Genotyping for HPV 16 / 18 may be valuable in patient management for predicting CIN / VAIN 2 / 3 in women with mildly abnormal Pap results.
PP 4: Performance characteristics of urinary tract Cytology: Observations from the College of American Pathologists interlaboratory comparison program in non-gynecological cytopathology (CAP NGC)
Guliz Barkan, MD, FIAC1, Manon Auger, MD2, Walid Khalbuss, MD, PhD3, Rodolfo Laucirica, MD4, Vijayalakshmi Padmanabhan, MD5, Rhona Souers, PhD6, Ann Moriarty, MD7
1Pathology, Loyola University, Maywood, Illinois; 2Pathology, McGill University, Montreal, Ontario, Canada; 3Pathology, University of Pittsburgh, Pittsburgh, Pennsylvania; 4Pathology, Baylor College of Medicine, Houston, Texas; 5Pathology, Dartmouth Medical School, Lebanon, New Hampshire; 6 Statistics, College of American Pathologists, Northfield, Illinois; 7Pathology, Ameripath Indiana, Indianapolis, Indiana
Introduction: Urinary tract cytology is currently used to evaluate patients with hematuria or as a management tool for those with urothelial neoplasia. This study investigates the performance characteristics of urinary tract specimens in the College of American Pathologists Educational Interlaboratory Comparison Program, CAP NGC, over an 11-year period.
Materials and Methods: The study evaluated participant responses between 2000 and 2010 from the CAP NGC with a reference diagnosis of ‘positive’ for malignancy (including high-grade urothelial carcinoma (HGUC), squamous cell carcinoma (SCC), or adenocarcinoma (ADC), and ‘benign’ diagnoses (such as Polyoma virus infection and ileal loop urine). The responses were analyzed with respect to the sample preparation type (conventional, liquid-based, and cytocentrifuged specimens) and participant type (laboratory, pathologist, cytotechnologist). The analysis was performed using a nonlinear mixed model, fitted with three factors — participant, preparation, and diagnosis types. The model also included the interaction terms between these factors and a repeated measures component to model the slide factor correlation structure. A P-value less than 0.05 was considered statistically significant.
Results: There were 96,093 responses from 1823 slides (46,637 pathologists, 29,976 cytotechnologists, 19,480 laboratories). Of the 74,821 responses for the ‘positive’ general diagnosis, 93.3% were concordant. Of the 21,272 responses for the ‘benign’ general diagnosis, 87.9% were concordant (P < 0.001). The main diagnostic difficulties in the malignant category were subtyping ADC and HGUC, and SCC versus HGUC. For the benign cases, the primary diagnostic pitfall was over-interpreting the negative cases, ileal loop specimens, and Polyoma virus specimens as HGUC. Within the positive reference diagnosis, cytotechnologists performed better than pathologists, whereas, pathologists performed better with the negative challenges (P < 0.001). Specifically, while pathologists were more likely to undercall HGUC as Polyoma, cytotechnologists were more likely to overcall Polyoma as HGUC (P < 0.001). Overall, the liquid-based samples performed significantly better than the other preparations (P < 0.001).
Conclusions: Urinary tract cytology preparations perform well in an interlaboratory comparison program. Liquid-based preparations performed the best. There were diagnostic difficulties subclassifying malignant cases as well as interpreting negative, ileal loop specimens, and Polyoma virus challenges as malignant.
PP 5: Malignant peritoneal and pleural fluid samples are adequate for molecular profiling
Raheela Ashfaq, MD, Yolanda Fong, MD
Caris Life Sciences, Irving, Texas
Introduction: Targeted therapy for cancer treatment is a growing aspect of clinical oncology and pathology. The Caris Target Now™ is a proprietary, evidence-based molecular profiling system for solid tumors providing specific and individualized molecular profiles for guidance of therapy in advanced stage and metastatic malignancies. The Target Now system is platform agnostic and utilizes data collected from a combination of immunohistochemical stains, Fluorescence in-situ hybridization (FISH), the Gene expression array, and sequencing tests, to give the final therapeutic guidance. A recent publication by Daniel Von Hoff et al. (JCO: 2010 Nov. 20; 28(33): 4877 – 83) shows improved progression-free survival with assay-guided therapy compared to physician-selected therapy. As acquisition of tissue in patients with advanced cancer can be challenging, we are reporting our experience with molecular profiling on malignant fluid samples.
Materials and Methods: A computer search was conducted to retrospectively identify malignant fluid samples or cell blocks submitted by various oncology service providers for the explicit purpose of molecular profiling of patients with advanced cancer. A cell block was either prepared or available for testing of all samples. A hematoxylin and eosin (H and E) slide was prepared from the cell block and reviewed by a pathologist before any testing. Malignant cell percentages were determined for the purpose of DNA microarray analysis and sequencing. Appropriate clusters and malignant cells were marked for FISH. The results were reviewed and data compiled, to calculate the yield of various molecular predictive tests.
Results: From January 2009 to April 2011, we studied 172 samples of peritoneal and pleural fluids. In order of frequency, the most common primary sites were, the lung (n = 49, 28.4%), ovary (n = 45, 26.1%), breast (n = 29, 16.8%), and pancreas (n = 5, 2.9%). The rest of the cases (44, 25.5%) included colon, peritoneum, endometrium, and others. We were able to perform more that 10 immunohistochemical stains in 123 samples (71.5%), 1 – 9 in three samples (1.7%), while 46 samples were insufficient for immunohistochemical analysis (26.7%). DNA microarray analysis was performed in 60 cases. FISH analysis was performed in 51 cases, and DNA sequencing for KRAS, EGFR or BRAF in 34 cases. The combined results of predictive markers from these various platforms were able to provide information on the therapeutic guidance for associated clinical benefit or lack of clinical benefit for various therapies in 129 of the 172 cases (75%).
Conclusions: Our study showed that cell blocks from malignant peritoneal and pleural fluid samples are informative and provide important therapeutic guidance in a large percentage of the cases. Molecular Profiling of malignant fluids offers opportunities for testing those patients where other tissue samples such as needle core biopsy or resection samples are not available.
PP 6: The optimal z-axis interval and focal planes to digitize 3-D gynecological SurePath® glass slides: Initial findings
Stanley Radio, MD1, Maheswari Mukherjee, MS, CT(ASCP)1, Najia Wright, MBA1, Jane Meza, PhD2, Amber Donnelly, PhD, MPH, SCT(ASCP)1
1Cytotechnology Education, School of Allied Health Professions, University of Nebraska Medical Center, Omaha, Nebraska; 2College of Public Health, University of Nebraska Medical Center, Omaha, Nebraska
Introduction: Virtual microscopy (VM) scans an entire glass slide and allows access to all areas of interest via a computer or digital device, without the use of a microscope. Application of VM to cytology has been dependent on the development of a three-dimensional focus. Our main objective is to determine the optimal 3-D scanning interval and focal planes necessary to achieve the best imaging resolution of the whole digitized SurePath® slide–prepared gynecological specimens, while maintaining a minimum file size. We are reporting the results of our pre-pilot study, which allows us to decrease the potential number of focal planes and interval levels.
Materials and Methods: [Table 1] Seventeen liquid-based preparations of gynecological (SurePath®) glass slides were scanned, each at 40X magnification: First, with 13 focal planes (FP) at 1 micron interval (Group 1), second, with 13 FP at 0.8 micron interval (Group 2), and finally with 13 FP at 0.5 micron interval (Group 3) using an iScanCoreo Au scanner (BioImagene, California, USA). Thus a total of 51 virtual images (VI) were produced with an average file size of 12.8 gigabytes. A cytopathologist, cytotechnologist, and cytotechnology student used Image viewer software and diagnosed the pre-annotated cells of interest in Groups 1, 2, and 3. They also recorded the FP level, with which they were confident in giving the diagnosis. Subsequently, they diagnosed the corresponding 17 glass slides (Group 4) using conventional microscopy. The participants were selected from these diagnostic categories: Negative, Negative with organism, Low-grade dysplasia, and High-grade dysplasia. We evaluated the interobserver reliability by using the kappa statistics. We also calculated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for all interval levels.
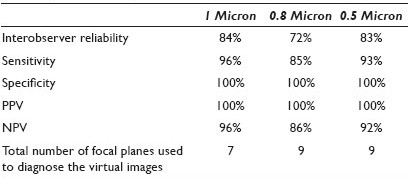
Results: The interobserver reliability was found to be the highest for the glass slides (100%) followed by VI scanned at 1 micron (84%), 0.5 micron (83%), and 0.8 micron (72%). When compared with the glass slides, the sensitivity was the highest for the VI scanned at the 1 micron interval (96%), followed by 0.5 (93%), and 0.8 (85%). The NPV was highest for the 1 micron interval (96%), followed by 0.5 (92%), and 0.8 (86%). The specificity and PPV was 100% for all interval levels. The number of focal planes used to diagnose the VI was seven for the VI scanned at 1 micron and nine for either 0.5 or 0.8 microns. One of the main comments from the participants was the difficulty in evaluating the chromatin pattern on dark cell clusters leading to inability to accurately interpret the virtual images.
Conclusions: Our study found that VI scanned at the 1 micron interval potentially had the highest interobserver reliability, sensitivity, and NPV, while using a lower number of focal planes. With this pre-pilot study the number of focal planes was decreased from thriteen to seven and the interval level established at 1 micron. This study will be followed up by a pilot study in which 20 SurePath® gynecological glass slides are scanned, each at: seven focal planes at 1 micron, five focal planes at 1 micron, and three focal planes at 1 micron. A sample size calculated from the pilot study will be used to determine the optimal scanning focal plane and the interval necessary to digitize SurePath® prepared gynecological specimens. In response to participants’ comments, an image enhancement option and an additional diagnostic category, ‘Unsatisfactory for Diagnosis’ will be added in the pilot study.
PP 7: Early clinical results on the implementation of ultrasound use in an academic fine needle aspiration service
Joseph Jakowski, MD, Celeste Powers, MD PhD, Alycia Reid, RT RDMS
Pathology, Virginia Commonwealth University Health Systems, Richmond, Virginia
Introduction: Ultrasound (US) has been successfully incorporated into the practice of many other medical subspecialties beyond radiology. In our pathology-based FNA service, we have seen superficial FNAs (SFNA) decrease substantially over the last five years in favor of image-guided FNAs. US and US fine needle aspiration (USFNA) are quickly gaining acceptance as a new tool for the interventional cytopathologist for real time evaluation and FNA of the target lesion. USFNA was recently incorporated into the SFNA service at our academic medical center and its initial impact and clinical utility are evaluated herein.
Materials and Methods: Retrospective evaluation of the first four months’ experience (12 / 17 / 2010 – 4 / 19 / 2011) of our new USFNA service was performed. Prior to initiation of the USFNA service, a discussion was held with the clinicians. Electronic templates (E and M, US, and FNA) and an image database were developed. All FNA requests were screened by the cytopathologist for appropriateness. Criteria for USFNA included: Non-palpable or ill-defined lesions, < 1.5 cm, > 25% cystic, any thyroid or breast mass, specific request, prior failed radiologically guided FNA, and / or masses near critical structures. Our cytopathologists are, at minimum, certified from the College of American Pathology in USFNA. Our ultrasonographer joined the service in month four and is certified by the American Registry of Diagnostic Sonography, with five years of US experience. US scans were performed using a high resolution US instrument with a linear transducer. All USFNA were performed by the cytopathologist with graduated training: Initially on turkey phantoms, then FNA of palpable lesions, and finally FNA of non-palpable masses. Patient evaluation included review of previous radiographic studies, a focused history and physical examination, and a US scan for echolocation.
Results: The SFNA service averaged 23 FNA requests per month (93 total) over the four-month study period. US use in the first three months of implementation of our new service varied from 0 to 25% of the cases per month and increased to 64% in month four, with the addition of a full-time ultrasonographer. In 71% of the patients, a USFNA was performed after echolocation of the lesion; 37.5% of these lesions were non-palpable, identified only on prior radiographic studies (diagnostic US or CT). The FNA sites included soft tissue (28%), breast (28%), thyroid (22%), and lymph nodes (22%). The overall specimen adequacy rate was 95.2%. In the remaining 29% of the patients, no USFNA was performed after an initial US scan for reasons that included: (1) The lesion was too deep, (2) it involved important anatomical structures (e.g., near lung apex), (3) there was a benign clinical and US impression such that follow-up was preferred, and (4) no discrete lesion was identified on US.
Conclusions: We have observed a number of immediate and encouraging consequences by incorporating US into our SFNA service. First, our SFNA service capabilities have been expanded to include aspiration of non-palpable lesions of superficial sites and second, patient care has been improved by appropriate elimination of some procedures. We have also had positive responses from our patients, pathology residents, and referring clinicians; the latter have increased their requests for this service. Finally, the addition of a full-time ultrasonographer has increased our use of US for evaluation, and not surprisingly, the number of USFNAs performed.
PP 8: Virtual microscopy in cytotechnology education: Application of knowledge from virtual to glass
Maheswari Mukherjee, CT(ASCP)1, Elizabeth Lyden, MS2, Amber Donnelly, PhD, MPH, SCT(ASCP)1
1Cytotechnology Education, School of Allied Health Professions, University of Nebraska Medical Center, Omaha, Nebraska; 2College of Public Health, University of Nebraska Medical Center, Omaha, Nebraska
Introduction: Virtual microscopy (VM) is a technology in which glass slides are scanned and converted into digital images. This study reports the implementation of VM in the cytotechnology educational program at the University of Nebraska Medical Center. The main objective of this study was to determine if cellular morphology learned through virtual microscopy could be applied to glass slide screening.
Materials and Methods: A total of 142 glass slides (61 teaching and 81 practice) of breast, thyroid, and lymph node fine needle aspiration body sites, were scanned with a single focal plane (at 40X) using iScan Coreo Au (BioImagene, California, USA). Six students, including one distant student, used only digital images to learn cellular morphology and conduct daily screening. Subsequently, the students were tested on 10 glass slides using light microscopy (LM). At the end of the study, the students were asked to respond to a survey, anonymously and voluntarily, about their VM experience. The survey had a series of questions followed by an open comment section for the students to give any additional feedback. The glass slide screening test scores of the participating students who learned through VM and tested on glass slides (VMLM group) were compared with the last three classes of students who learned through LM and tested on glass slides (LMLM group).
Results: A non-parametric statistical analysis indicated no difference (P = 0.14) in the median test scores between VMLM (median = 94) and LMLM (median = 86) groups. The survey indicated that the students spent a longer time screening virtual images compared to their previous experience of glass slide screening. They strongly believed that faculty member guidance and interaction would have improved the outcome of their virtual slide screening. In general, the students preferred LM over VM. However, the annotated teaching slides and access to the VM slides off campus were well appreciated by the students.
Conclusions: Although the students preferred LM, they were able to apply the cytological criteria learned through VM to glass slide screening. One of the main reasons for their preference for LM was that it took less time to screen the glass slides than the VM images. Overall, VM was considered a great teaching tool in conjunction with LM, but not to be used as the sole method of instruction or for daily screening of practice slides.
PP 9: Validation of BRAF mutational analysis in indeterminate thyroid fine needle aspirations
K. Councilman, MD1, N. Thomas, BS1, J. Bohn, BS1, P. Chesnut, BS1, B. Haugen, MD2, J. Klopper, MD2, M. Said, MD, PhD1, W. Franklin, MD1, D. Aisner, MD, PhD1
1Pathology, University of Colorado Anschutz Medical Campus, Aurora, Colorado; 2Endocrinology, University of Colorado Anschutz Medical Campus, Aurora, Colorado
Introduction: Multiple studies have demonstrated the utility of BRAF V600E mutational analysis for refining the diagnosis of indeterminate thyroid lesions. In addition, some studies suggest that mutation testing can aid in the stratification of patients with papillary thyroid carcinoma (PTC) diagnosed by fine needle aspiration (FNA). There are multiple methodologies for sampling thyroid lesions for molecular testing and performing mutational testing on thyroid FNAs. Methods for mutational testing should either be sufficiently sensitive to detect mutation despite a significant population of non-lesional cells, or involve enrichment for lesional cells. Here we present the technical validation of an approach that utilizes the microdissection of lesional cells directly from Diff-Quik® stained slides followed by conventional sequencing for BRAF V600E mutation.
Materials and Methods: Thirty seven thyroid FNA cases were identified based on a retrospective review of the pathology information system. Eleven cases were diagnosed as negative for malignancy, nine were diagnosed as malignant and were all classified as PTC, and 17 cases were classified as ‘indeterminate’ based on the recent Bethesda classification system, including diagnoses of ‘atypical cells’, ‘suspicious for malignancy’, ‘follicular lesion,’ and ‘follicular neoplasm’. Criteria for testing were, (1) minimum of one cluster of at least 10 lesional cells on a Diff-Quik® slide and (2) diagnostic material remaining in the archive if the identified Diff-Quik® slide was sacrificed for testing. Lesional cells were selectively microdissected from de-coverslipped Diff-Quik® slides under a dissecting microscope and DNA was extracted from the microdissected material. Direct sequencing of exon 15 of BRAF was performed to evaluate for the presence of BRAF V600E mutation. Of the 37 cases selected, 16 had corresponding follow-up resection material, which was also evaluated for the presence of BRAF V600E mutation.
Results: All cases yielded sufficient DNA to undertake mutational testing. Of the 37 cases, five had BRAF V600E mutations, all of which were diagnosed as PTC on cytology. None of the 11 negative or 17 indeterminate cases demonstrated a BRAF V600E mutation. Paired evaluation of the cytology specimen and follow-up resection specimen showed concordance in all cases except one, which indicated the absence of a mutation in the cytology specimen, but positive mutation status in the follow-up resection specimen. Additional testing of the surgical resection specimen demonstrated areas that were negative for mutation, suggesting heterogeneity for mutation within the tumor.
Conclusions: A high correlation between the BRAF mutation status and diagnosis was observed. Furthermore, a high correlation between the BRAF status on FNA and the subsequent resection specimen was seen. These findings indicate that selective microdissection of Diff-Quik® slides, followed by direct sequencing is an effective method to evaluate the BRAF mutational status of thyroid lesions. Although no mutation was identified in the indeterminate lesions tested, this finding is not unexpected, as several studies have required a larger number of indeterminate specimens in order to identify BRAF mutations. These findings further suggest that BRAF testing can be performed retrospectively, and does not typically require additional sampling or FNA passes to obtain material for testing.
PP 10: Assessment of Fine Needle Aspiration specimen adequacy for high risk HPV detection and genotyping in oropharyngeal squamous cell carcinoma
Charalambos Solomides, MD, Marluce Bibbo, MD, Zi-Xuan Wang, PhD
Pathology Anatomy and Cell Biology, Thomas Jefferson University Hospital, Philadelphia, Pennsylvania
Introduction: In patients with oropharyngeal squamous cell carcinoma (SCC), the presence of human papillomavirus (HPV) genotype 16 is a favorable prognostic indicator, with respect to recurrence and overall survival. Thus, fine needle aspirates (FNA) of these tumors should be properly handled to provide both morphological and molecular information. The aim of this study was to determine the adequacy of the archived and fresh FNA specimens for the molecular detection and genotyping of HPV.
Materials and Methods: Sample selection: A total of 37 specimens from 26 patients diagnosed with metastatic SCC during the last five years were available in the cytology laboratory of the TJU hospital to be included as retrospective specimens. Among these specimens, 19 had slides stained with Papanicolaou (Pap) and 18 with Diff-Quik® (DQ). Nine fresh FNA specimens from nine patients with metastatic SCC diagnosed during the last two months were included as prospective specimens. HPV analysis: Pap or DQ stained slides were soaked in xylene overnight to remove the cover slips. Cells on these slides were scraped into tubes, washed with ethanol, and dry pellets were resuspended in 2 ml of PreservCyt, a liquid-based cytology solution. The needles containing fresh residual FNA specimens were directly rinsed in 2 ml of PreservCyt solution. These specimens were then analyzed using the standard protocol for ThinPrep® cervical specimens, with the Cervista HR HPV detection kit. The positive specimens were then tested for the HPV 16 and 18 genotypes.
Results: The Cervista HR HPV detection assay used a housekeeping gene (human histone HIST2H2BE gene) as an internal control to assess if there were sufficient cells in the specimens for the detection of HR HPV. Overall, 36 of 46 specimens (78%) had sufficient cells to yield a valid HPV result. The adequacy rate for Pap-stained slides was 14 / 19 (74%), for DQ stained slides 14 / 18 (78%), and 8 / 9 (89%) for fresh needle aspirates. HR HPV was detected in 17 specimens. Identical HR HPV results were obtained from parallel specimens stained with DQ and Pap stains in six patients. Among the 17 HPV-positive specimens, 12 were genotyped as HPV 16, two were HPV 18, two contained both HPV 16 and 18. One specimen was unavailable for genotype analysis.
Conclusions: In our preliminary experience, HPV 16 detection and genotyping could be performed on FNA specimens prospectively collected in PreservCyt as well as archived slides stained with either Pap or DQ. This pilot study demonstrated the clinical utility of genotype testing for high-risk HPV strains when cytopathology materials, either fresh or archived, were available for analysis.
PP 11: Using Fluorescence in-situ hybridization and polymerase chain reaction in the diagnosis and classification of lymphoproliferative disorders on fna material: Eight-year experience from a medical center
Xiaohong Wang, MD, PhD, Fleurette Abreo, MD, Jaiyeola Thomas, MD, Diana Veillon, MD, James Cotelingam, MD, Songlin Zhang, MD, PhD
Pathology, LSUHSC, Shreveport, Louisiana
Introduction: The role of fine needle aspiration in the assessment of lymphoproliferative disorders continues to be a subject of debate. Many studies have shown the accurate diagnosis and classification of lymphoma using FNA. Increasing publications in the past few years have shown the value of Fluorescence In-Situ Hybridization (FISH) using cytology preparation in the classification of lymphoproliferative disorders on FNA. Previously, we published two years of our experience and our algorithm for handling lymphoproliferative disorders on FNA. Here, we summarize the past eight years of our experience using FISH and Polymerase Chain Reaction (PCR) for the diagnosis and classification of lymphoma using FNA material.
Materials and Methods: A retrospective search from the pathology data base for FISH and PCR on FNA materials in the past eight years (2003 and 2010) was performed, and the results of the corresponding cytology diagnosis and flow cytometry results were reviewed.
Results: A total of 53 FNA cases had FISH and / or PCR testing during the study period, and the cytology / flow cytometry diagnoses included 32 lymphomas, 16 atypical, and five negative. Molecular tests included 33 FISH for targeted translocations, 11 FISH IgH, five PCR for B-cell clonality, and eight PCR for T-cell clonality. The 32 lymphomas were further classified as 12 follicular lymphomas, eight diffuse large B-cell lymphomas (DLBCL)-NOS, four Burkitt lymphomas, one peripheral T-cell lymphoma, one plasmablastic lymphoma, one indolent B-cell lymphoma, and five B-cell lymphomas without classification. The 16 atypical cases were further classified as two peripheral T-cell lymphomas, one MALToma, two DLBCL-NOS, two thymomas, and nine atypical / suspicious. The five negatives were further defined as one atypical and four negatives. Twenty-five cases had follow-up excisional biopsy, including 21 lymphomas, two thymomas, and two negatives. There were no false positive or negative cases based on the cytology / flow cytometry / molecular interpretation. For individual methods, there were four false negatives on flow cytometry, four false negatives on FISH IgH, and one false negative on PCR. All 16 lymphomas, with sub-classification, were confirmed on excisional biopsy. The remaining five cases were atypical diagnoses on cytology / flow cytometry and they could not be further defined or classified with FISH / PCR. The final surgical diagnoses of the five cases were three DLBCL, one nodal marginal zone lymphoma, and one Hodgkin lymphoma.
Conclusions: FNA cytology combined with flow cytometry immunophenotyping and targeted FISH on FNA material could accurately diagnose and sub-classify most of the non-Hodgkin lymphomas. Targeted FISH and PCR could further diagnose and then sub-classify atypical / suspicious cases. Due to the complexity of non-Hodgkin lymphoma classification, using the appropriate algorithm with a team approach, including hematopathologists, is necessary to render an accurate diagnosis with a useful classification. Excisional biopsy required on some difficult cases should be performed without hesitation; however, the significant value of FNA cannot be underestimated.
PP 12: Detection of cytogenetically abnormal circulating cells in lung cancer patients in peripheral blood using an antigen-independent fluorescence in-situ hybridization (FISH) assay: A case control validation study
Jeff Wang1, Tanweer Zaidi1, Weigong He1, Jieqing Chen1, Pathak Aditya1, Jeena Vaid1, Margaret Spitz2, Carol Etzel2, Randa El-Zein2, Ruth Katz1
1Pathology Department, MD Anderson Cancer Center, Houston, Texas; 2Epidemiology, MD Anderson Cancer Center, Houston, Texas
Introduction: Detection of circulating tumor cells using a simple blood test may provide a minimally invasive method to assist in early diagnosis of indeterminate lung nodules representing lung cancer (LC) and to monitor the results of the therapy. In a pilot study, using 12 DNA probes known to be affected in LC, we previously demonstrated via an automated FISH scanner that an antigen-independent FISH assay was a sensitive and quantitative method to detect cytogenetically abnormal cells (CACs) in the peripheral blood (PB) of non-small cell lung cancer patients (NSCLC)[1]. The different DNA probe sets could distinguish cancer patients from controls, as also the number of CACs correlated with the stage of disease, with early stage patients showing a lower number of CACs than the advanced-stage patients[1]. In contrast to the existing methods of using EpCAM-coated beads to isolate circulating tumor cells, our assay showed much higher numbers of CACs than previously reported. The current study was performed on a new cohort of LC patients and controls, to validate if by using only two DNA probe sets [centrometric 10 / 10q22.3 (SP-A) and centromeric 3 / 3p22.1 (GC20)], we could still detect lung cancer in the PB samples.
Materials and Methods: The PB samples from consented LC patients [adenocarcinoma (12), squamous cell carcinoma (six), small cell carcinoma (two) and carcinosarcoma (one)] and 18 low and high-risk controls were evaluated. The demographic data (age, smoking history, and cancer status) was anonymized so that reviewers of the FISH images were blinded. PB mononuclear cells (PBMCs) were isolated from 10 ml of PB and hybridized with two, two-color (3p22.1 / CEP3 and CEP10 / 10q22.3) FISH probes and quantified on an automated fluorescence scanner (Bioview Duet™, Il). For each sample, chromosomal abnormalities in 500 intact round or oval cells were classified by one reader and confirmed by a second reader as monosomies (mono), polysomies (poly) loss / gain, of 3p22.1 / CEP3 and 10q22.3 / CEP10, as previously described (1). All abnormalities (abn) of 3p and 10q were summed. CACs in PB were calculated (% chromosomal abnormalities X total PBMC / ml PB) / 1000), and expressed as CACs per microliter of blood. The mean ± S.D. of all chromosomal abnormalities and CACs were analyzed by the Kruskall Wallis test for significance (P < 0.05). A total of 39 samples (21 patients and 18 controls) were evaluated.
Results: Compared to the control group, the LC group showed significantly higher percentages of CACs with del 3p22.1 and 10q22.3 (P < 0.001,0.002), and abn 3p22.1 and 10q22.3 (P < 0.001, P < 0.001); poly, gain, abn of 10q22.3 (P = 0.005, P = 0.004, P = 0.007); mono, poly, gain, and abn 3p22.1 + 10q22.3 (P = 0.04, P = 0.000, P = 0.001, P = 0.002); poly, gain, and abn3p22.1 (P = 0.004, P = 0.003, P = 0.008).
Conclusions: In a validation study, comprised of 39 blinded PB samples, we have demonstrated that it is possible to detect LC using an automated FISH test, which uses four DNA probes known to be important in the pathogenesis of LC. We showed highly significant correlations between the numbers of CACs as well as percentages of chromosomal abnormalities in LC patients compared to controls. We plan to accrue larger numbers of samples.
PP 13: Accuracy of the cytology specimen and needle core biopsies for detection of KRAS mutation in non-small cell carcinoma: Comparison with resection specimen
Ismatun Swati, MD, Shengle Zhang, MD, Jamie Tull, MS, Kamal Khurana, MD
Department of Pathology, SUNY Upstate Medical University, Syracuse, New York
Introduction: KRAS mutational analysis is critical for predicting the anti-EGFR therapeutic response in non-small cell lung carcinoma (NSCLC). Needle core biopsy and a cytology specimen from endobronchial ultrasound-guided transbronchial needle aspiration as well as CT-guided fine needle aspiration (FNA) play an important role in the diagnosis and staging of NSCLC. However, the concordance rate of KRAS mutation analysis between the cytology specimen / needle core biopsy and resected NSCLC is unknown. We evaluated the extent to which the KRAS mutation detection of a cytology specimen or needle core biopsy of primary lung tumor by comparing the results with the resected NSCLC from the same patient.
Materials and Methods: Twenty-five samples including eight cell blocks, seven cytology smears, and 10 needle core biopsies, and the corresponding 22 resection specimens of the primary lung tumor were correlated for KRAS mutational analysis. Formalin-fixed paraffin embedded cell blocks, needle core biopsies, resected tumor sections, and alcohol-fixed smears were used for isolation and amplification of DNA. Direct sequencing was applied to detect KRAS mutations in codon 12 and 13. In cases where the cell block material did not correspond with the results on the resected specimens, cytology smears of the corresponding cases were used for isolation of DNA, using microdissection.
Results: Sufficient DNA for PCR was successfully isolated on all cytology and surgical (biopsies and resection) specimens except for one cell block. KRAS mutation was detected in seven out of 10 needle core biopsy specimens and the corresponding surgically resected specimens, with 100% concordant results. KRAS mutation was detected in four out of eight cell blocks, compared to seven out of the eight corresponding surgically resected samples. Lack of correlation in three cases with cell blocks could be attributed to low cellularity (two cases) and failure to retrieve DNA (one case). However, use of cytology smears in these three cases confirmed the KRAS mutation noted in the corresponding surgically resected samples. In addition, four more smear samples were tested and showed 100% concordance with the corresponding surgically resected samples.
Conclusions: KRAS mutation analysis on the cytology specimen and needle core biopsies is highly accurate and comparable with that of the resected NSCLC. In cases with low cellularity on cell blocks, cytology smears provide a better substitute for KRAS mutation analysis, when using the microdissection technique.
PP 14: Identification of epidermal growth factor receptor mutation, K-ras mutation, and EML4-ALK translocation in the cytological specimens of primary and metastatic non-small cell lung cancers
Guoping Cai1, Gillian Levy1, David Chhieng1, Robecca Wong1, Scott Gettinger2, Jonathan Puchalski2, Robert Homer1, Pei Hui1
1Pathology, Yale University School of Medicine, New Haven, Connecticut; 2Internal Medicine, Yale University School of Medicine, New Haven, Connecticut
Introduction: Although its carcinogenesis is not fully understood, non-small cell lung cancer (NSCLC) has been shown to be associated with certain molecular alterations. Identification of molecular changes such as epidermal growth factor receptor (EGFR) mutation, K-ras mutation, and EML4-ALK translocation has therapeutic implications in the clinical management of NSCLCs. In case of lung cancers, the cytological specimens from pleural effusion or fine needle aspirates might be the only material available for molecular analysis. However, some factors encountered in the cytological preparations such as low cellularity may have a potential impact on the final outcome of the molecular tests. This study is to review our experience with molecular tests in the cytological specimens of primary or metastatic NSCLCs.
Materials and Methods: A total of 55 cases of NSCLCs that had molecular analysis were retrieved from the cytopathology archives of our institution. There were 12 primary and 43 metastatic NSCLCs, including 50 cases of adenocarcinomas. The metastatic sites included pleural / pericardial effusion (19 cases), lymph node (16 cases), soft tissue (3 cases), bone (2 cases), adrenal gland (2 cases), and liver (1 case). Molecular tests were performed on the cell-block materials of effusions (19 cases) or fine needle aspirates (36 cases). EGFR mutation was evaluated by PCR-sequencing analysis of exons 18, 19, 20, and 21. K-ras mutation was tested using PCR-single strand conformational polymorphism analysis of codons 12 and 13. EML4-ALK translocation was evaluated by FISH study using the Anaplastic Lymphoma Kinase (ALK) break-apart probe.
Results: The patients were 25 males and 30 females with ages ranging from 28 to 90 years (average 68). Molecular tests were successful in 50 of 55 cases (91%). The remaining five cases yielded no results due to insufficient material. Evaluation of EGFR mutation, K-ras mutation, and EML4-ALK translocation were performed in 42, 14, and 22 cases, respectively. EGFR mutations were found in 14 of the 42 cases (33%), including deletion mutation in exon 19 (six cases), L858R mutation in exon 21 (five cases), and T790M mutation in exon 20 (three cases). K-ras point mutation was detected in three of 14 cases (21%) including TGT mutation in codon 12 in two cases and AGT mutation in codon 12 in one case. EML4-ALK translocation was evident in three of the 22 cases (14%). All the molecular alterations detected were mutually exclusive for the cases tested. Six of the twelve primary tumors showed EGFR (five cases) or K-ras mutation (one case). EGFR mutation, K-ras mutation, and ELM4-ALK translocation were identified in nine, two, and three of the 38 metastatic tumors, respectively.
Conclusions: The current study demonstrates the feasibility of molecular tests in the cytological specimens of both primary and metastatic NSCLCs. The presence of EGFR mutation, K-ras mutation or EML4-ALK translocation, appears to be mutually exclusive in NSCLCs.








