Translate this page into:
Abstracts for the 59th Annual Scientific Meeting (November 2011) by American Society of Cytopathology (ASC) at Baltimore, MD, USA
-
Received: ,
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Medknow Publications and was migrated to Scientific Scholar after the change of Publisher.
Abstract
These are peer-reviewed poster-platform submissions finalized by the Scientific Program Committee. A total of 153 abstracts (14 Platforms [PP1 through PP14] & 139 Posters [1 through 139]) were selected from 161 submissions to be considered for presentation during November 4 – 8, 2011, at the Hilton Baltimore Hotel, to pathologists, cytopathologists, cytotechnologists, residents, fellows, students, and other members of cytopathology-related medical and scientific fields.
Keywords
Abstracts
American society of cytopathology
ASC
cytopathology
cytology
127: Withdrawn
128: Discordances in immunoperoxidase stains performed in evaluation of carcinomas: A comparison between results on cytological and surgical specimens
Nyasha Bullock, MD, Mary Fiel-Gan, MD, Richard Cartun, PhD, Srinivas Mandavilli, MD
Pathology and Laboratory Medicine, Hartford Hospital, Hartford, Connecticut
Introduction: Immunoperoxidase (IPOX) stains are established ancillary methods for the evaluation of carcinomas on cytological specimens. IPOX stains are optimally performed on formalin-fixed, paraffin-embedded (FFPE) tissues, including cell blocks. However, FFPE tissue / cell blocks are not available in all cases and in such instances IPOX stains could be applied directly on alcohol-fixed cytological smears (AFCS). There is limited literature evaluating the reliability of the results of IPOX on AFCS. The aim of this study was to compare the results of IPOX stains performed on AFCS and FFPE tissue using the same cohort of cases.
Materials and Methods: Five hundred and eighty two tumors (primary and metastatic carcinomas from a variety of body sites) were retrieved from the pathology files over a two-year period (2008 – 2009) and 226 of those cases had a tissue follow-up. There were 57 cases in which IPOX stains were performed in both AFCS and surgical specimens. IPOX concordance (results reported as positive or negative in both specimens) or discordance (positive in one and negative in the other specimen) were tabulated. IPOX was performed on AFCS and FFPE using polymer-based HRP / DAB detection with careful evaluation of controls.
Results: There was 100% concordance in the following antibodies (numbers in parentheses represent # of cases): CK-7 (7), CK34BE12 (4), Calcitonin (4), Calretinin (3), Thyroglobulin (3), CA125 (2), CEA-m (2), CD117 (2), PLAP (1), Mammaglobin (1), PSA (1), CD34 (1), CD31 (1), PSAP (1), and WT1 (1). There was 100% discordance in the following antibodies: CD10 (1), EMA (1), MOC31 (1), Beta catenin (1); 50% discordance in the following antibodies: Hep Par1 (2), RCC (2); and 33% or less discordance with TTF-1 (13), CK cocktail (12), Synaptophysin (11), Chromogranin (11), ER (8), CK-20 (7), CDX2 (4), p63 (4), S-100 (4), and PR (3). The discordance was overwhelmingly represented in cases with IPOX negative on AFCS and IPOX positive FFPE material. There were only four cases (ER, p63, TTF-1 and RCC — all one each) that were IPOX positive on AFCS and negative on FFPE material.
Conclusions: (1) ICC stains can be successfully performed on AFCS when FFPE is not available, with 100% concordance seen in several commonly utilized antibodies. (2) Although this is a relatively limited study, the results suggest that there can be discordances in the IPOX results between AFCS and FFPE material and the vast majority of such discrepancies represent a likely ‘false negative’ IPOX result on AFCS. (3) Immunoperoxidase results have to be carefully interpreted on AFCS and evaluated in the individual clinical context.
129: Molecular testing in cytopathology
Frances Cate, MD, Doha Itani, MD, Cindy Vnencak-Jones, PhD, Joyce Johnson, MD, Alice Coogan, MD
Pathology, Vanderbilt University Medical Center, Nashville, Tennessee
Introduction: Molecular testing has widely become the standard of care for lung cancer and melanoma. Cytological samples are frequently the diagnostic material obtained due to decreased morbidity of the procedure, as compared to more invasive procedures. In July, 2010, molecular assays for melanoma and lung cancer became available at Vanderbilt University Medical Center (VUMC). This is a report of our laboratory's experience using cytology specimens for these analyses.
Materials and Methods: All tumors were tested in the Molecular Diagnostics Laboratory at VUMC using the ABI PRISM® SNaPshot® Multiplex Kit and a laboratory developed test, with appropriate positive and negative controls. The assay included testing for KRAS, BRAF, NRAS, PIK3CA, MEK1, AKT1, PTEN, and EGFR mutations in primary lung tumors and BRAF, NRAS, GNA11, GNAQ, KIT, and CTNNB1 mutations in melanomas. Routine molecular testing of cytology specimens was not implemented, but rather was performed only on request by the treating physician. The specimens were tested using a cell block or a spun-down fresh cell pellet. On cell blocks, the amount of submitted tissue was measured in millimeters. A percentage of the tumor cells present was determined by a pathologist, to establish the adequacy of the sample and judge whether it was possible to enrich for tumor. Our procedure was to enrich for tumor as much as possible. Samples having 10% or more tumor cells were considered adequate for testing, based on the sensitivity data established in the molecular diagnostics laboratory. For tissues that could not be enriched, the percentage of tumor cells was noted and paraffin sections were obtained. As for cases that could be enriched, unstained slides were obtained and macro-dissection of the enriched tumor areas was conducted, to acquire material for molecular testing. An H and E stained slide was prepared after tissue sectioning, to ensure that the tumor was consistently present in the intervening slides tested. Fresh tissue pellets were also accepted for testing provided a cytospin obtained from the same material met the adequacy criteria.
Results: From July 1, 2010 to February 28, 2011, 24 cytology specimens were referred for testing. Four had less than 10% tumor cells and were considered inadequate. One sample was referred to the laboratory in error and in one case, testing was canceled by the treating physician. The 18 remaining specimens included 15 lung cancers and three melanomas. Two of the lung cancers were tested with pellets of fresh tissue. The cell blocks on the remaining 16 samples had a range of 3 mm to 15 mm of tissue that was macro-dissected. Fifteen of these had greater than 10% tumor cells on the initial and final H and E slides. One sample of a primary lung tumor had 10% tumor cells on the initial H and E and 3% on the final H and E; however, the sample was tested at the request of the physician and a mutation was detected. Ten out fifteen lung tumors were positive for genetic mutations: Seven KRAS, two EGFR, and one MEK1. Nine mutations were identified from the cell block samples and one from a pellet sample. Two out of three melanomas were positive for NRAS mutations.
Conclusions: We conclude that the DNA yield from cytology specimens is adequate for molecular mutation analysis. Material submitted for testing should be enriched for tumor as much as possible in order to increase the confidence in targeted tumor testing. Specimens without a cell block can have reliable studies performed from a pellet of fresh cells.
130: A comparative study of thyroid ultrasound-guided fine needle aspiration biopsies performed by cytopathologists versus others
YoonSun Choi, CT(ASCP), David Burstein, MD, Hua Chen, MD, PhD, Hui Liu, MD, Sambasivarao Sunkara, SCT(ASCP), Arnold Szporn, MD, David Zhang, MD, PhD, Maoxin Wu, MD, PhD
Anatomic Pathology, Mount Sinai Medical Center, New York, New York
Introduction: In the past, the majority of thyroid fine needle aspirations (FNA) were done by palpation. Today there is an increased number of FNA biopsies performed routinely using ultrasound guidance (USG). The objective of this study was to evaluate the effectiveness of thyroid USG-FNA performed by pathologists compared with others, including endocrinologists and radiologists, from the institution.
Materials and Methods: Thyroid USG-FNA cases submitted during the period of September 1, 2010 to April 6, 2011 were reviewed. There were 134 performed by cytopathologists and 368 performed by others. The Bethesda System (BS) for thyroid cytology was applied to classify each group. A surgical follow-up study was also recorded. Statistical analysis for cases with surgical follow-up (SFU) was performed by using surgical diagnosis as a gold standard.
Results: As shown in Table 1, among the 134 cases performed by cytopathologists, there were four (3%) non-diagnostic / unsatisfactory (BS 1), 89 (66%) benign (BS 2), 11 (8%) atypical / follicular lesions of undetermined significance (BS 3), nine (7%) follicular neoplasms (FN) / suspicious for FN (BS 4), seven (5%) suspicious for malignancy (BS 5), and 14 (11%) malignant (BS 6), and there were 27 (20%) cases with SFU. In comparison, the 368 cases performed by others showed 76 (21%) BS 1, 238 (65%) BS 2, 26 (7%) BS 3, 10 (3%) BS4, nine (2.5%) BS 5, and nine (2.5%) BS6, and there were 26 (7%) cases with SFU. The statistical results based on SFU are presented in Tables 2 and 3.
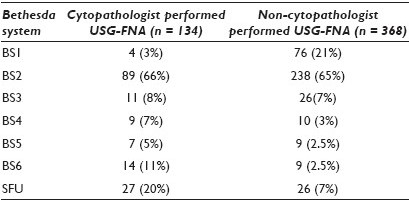
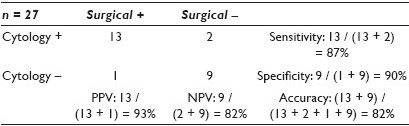
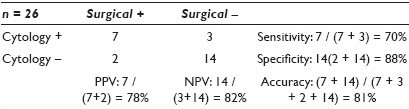
Conclusions: USG-FNA performed by pathologists with the onsite evaluated group shows less unsatisfactory cases (3% vs. 21%), higher rate of SFU (20% vs. 7%), and better sensitivity (87% vs. 70%), specificity (90% vs. 88%), and PPV (93% vs. 78%) as compared to the non-cytopathologist group.
131: Electromagnetic navigation bronchoscopy-guided fine needle aspiration for the diagnosis of lung lesions
Shelley Redfern, MD1, Thomas Gildea, MD2, Deborah Chute, MD1
1Pathology and Laboratory Medicine Institute, 2Respiratory Institute, Cleveland Clinic, Cleveland, Ohio
Introduction: Smaller lung nodules are being detected earlier due to the increasing use of improved imaging. Many peripheral lesions are beyond the reach of conventional bronchoscopes, and require CT-guided or open surgical biopsy, which carry increased risks to the patient. Electromagnetic navigation bronchoscopy (ENB) is a new technique, which uses an image-guided localization system to guide the bronchoscopic tools to predetermined points, within the bronchial tree. Preliminary studies have demonstrated the safety and feasibility of this technology. In this study, we investigate the sensitivity and specificity of ENB-guided fine needle aspiration (FNA) in the diagnosis of lung lesions.
Materials and Methods: All ENB-guided FNAs performed at one institution were included in the study. The superDimension / bronchus system (superDimension Inc, Plymouth, Michigan, USA) was used in all cases. The pathological reports of the ENB-guided FNAs, as well as all the other pathological material, obtained within an 18-month period, were reviewed. Patients with a positive ENB-guided FNA or malignancy within the same lobe were considered positive for malignancy. Patients with an atypical diagnosis but no definitive malignancy were considered negative for malignancy, for statistical purposes. Histological diagnosis was considered the gold standard when available.
Results: Ninety-one patients underwent 95 ENB-guided FNAs over a three-year period (32 RUL, 8 RML, 14 RLL, 28 LUL, 2 lingula, and 11 LLL). Thirty-four patients (37%) were positive for malignancy during the study period (14 adenocarcinoma, 9 squamous cell carcinoma, 7 non-small cell carcinoma NOS, 1 small cell carcinoma, 3 other). ENB had a sensitivity of 65% and a specificity of 86% for the detection of malignancy [Table 1]. Of the patients who were negative by ENB-guided FNA and had confirmed malignancy by another technique — four were positive on concurrent bronchial biopsy, four were positive on concurrent bronchial brushings, and two were positive on subsequent FNA. Of the patients negative for malignancy on all procedures — seven patients (12%) had findings consistent with an infectious process by at least one procedure (1 aspergillus, 6 granulomatous inflammation). ENB-guided FNA identified the infectious process in three patients (43%).

Conclusions: Electromagnetic navigation bronchoscopy is a new technique that is not widely used yet. In our institution, ENB-guided FNA has a sensitivity of 65% and specificity of 86% for the diagnosis of malignancy. The overall diagnostic yield from any procedure performed at the time of ENB has been 74%. ENB-guided FNA demonstrates good utility for the diagnosis of difficult-to-access lung lesions, especially when used in conjunction with other techniques.
132: Clinical experience in the application of a novel mRNA-based gene expression classifier in thyroid nodules with indeterminate fine needle aspiration cytology
Robert Monroe, MD1, Richard Lanman, MD1, Giulia Kennedy, PhD1, Jonathan Romanowsky, MBA1, Ken Brunt, BS1, Katie O’Reilly, MD2, Carola Zalles, MD2, S. Thomas Traweek, MD2
1Veracyte, Inc., South San Francisco, California; 2Thyroid Cytopathology Partners, Austin, Texas
Introduction: The Afirma® Gene Expression Classifier is a multi-gene expression test for the analysis of fine needle aspirates (FNA) from thyroid nodules, with indeterminate cytopathology. Postoperatively, two out o three of these indeterminate nodules were diagnosed as benign. Preoperatively, the test identified indeterminate nodules that were benign, to avoid diagnostic thyroid surgery. However, performance of the test in clinical practice has not yet been reported.
Materials and Methods: Diagnoses were made by a panel of cytopathologists experienced in interpretation of slides utilizing liquid-based preparation as well as direct smears, and classified them in accordance with the bethesda system for reporting thyroid cytopathology. Indeterminate cytopathology included atypia or follicular cells of undetermined significance (AUS / FLUS), follicular and Hürthle cell neoplasm or suspicious for same (FN / SFN), and suspicious for malignancy. Reflex molecular testing was performed on FNAs with indeterminate cytopathology, utilizing the mRNA expression of 142 genes and a proprietary algorithm, as previously described.
Results: The first 1,025 consecutive FNA samples were submitted by 62 physicians practicing in academic (9%) and community settings (91%). The cytopathologists reviewed 928 FNAs and reported 70.5% as benign, 0.4% as malignant, 15.4% as indeterminate, and 13.7% as nondiagnostic. Ninety-seven FNAs were submitted directly to the gene expression classifier. The gene expression classifier results on the 226 indeterminate samples were: 53% benign and 38% suspicious (meaning not definitively benign), and 9% ‘no result’, [Figure 1]. All the ‘no result’ samples were AUS / FLUS. One hundred and twenty-four FNA samples with indeterminate subtype were classified in accordance with Bethesda by the cytopathology panel. Of these, 25% of the cytological suspicious for malignancy were molecular benign, as were 56% of the FN / SFN and 53% of the AUS / FLUS, [Table 1].
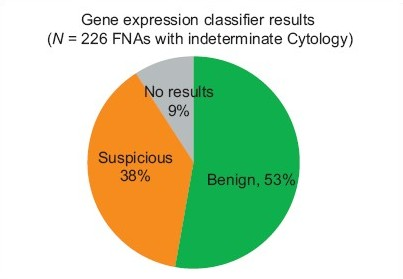
- Gene expression classifier results on 226 fnas with indeterminate cytopathology
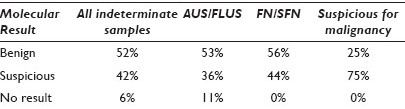
Conclusions: The benign, indeterminate, and nondiagnostic rates we report are consistent with the literature, however, our cytology malignant rate is lower. Fifty percent of the nondiagnostic samples were submitted by eight of 62 physicians (13%), suggesting that the nondiagnostic rates may be related to FNA technical skill or selection bias. There was little difference in the cytology subtype AUS / FLUS and FN / SFN by molecular analysis, which had molecular benign results of 53 and 56%, respectively. The cytology subtype suspicious for malignancy was benign molecularly 25% of the time, somewhat lower than the 38% benign found postoperatively in a recent meta-review. On the basis of the test specificity reported in two prospective validation studies, and the risk of malignancy in cytologically indeterminate nodules (34%), one would expect the molecular result to be benign in approximately 50% of the cases. This projection has now been accurately reflected in actual experience. A comparison of the rate of benign results by molecular analysis found slightly higher rates at the academic sites versus community-based sites, but the difference was not statistically significant. The molecular test has the potential to reduce the number of diagnostic thyroidectomies on nodules, with indeterminate FNA cytology, by more than 50%.
133: Identification of tumor proteins associated with early stage of lung adenocarcinoma by quantitative proteomics
Qing Li, MD, PhD, Yan Li, PhD, Frederic Askin, MD, Edward Gabrielson, MD, Hui Zhang, PhD
Pathology, The Johns Hopkins Medical Institutions, Baltimore, Maryland
Introduction: Lung cancer is one of the leading causes of cancer-related deaths worldwide, and adenocarcinoma is now the most common histological variant. Although the targeted therapies are progressed rapidly in recent years, based on the discovery of molecular markers, the early detection of lung cancer and identification of prognostic biomarkers are still suboptimal. Over 60% of lung cancer patients present as locally advanced or metastatic disease at the time of diagnosis; and a progression-free survival rate is markedly different even between stage I and stage II cancers. Few previous studies have focused on the identification of tumor proteins in the early stage of lung cancer. In this study, we quatitatively analyzed and compared tumor proteins in both stage I and II lung adenocarcinomas using novel quantitative proteomics.
Materials and Methods: Paraffin-embedded lung adenocarcinoma tissues were collected and tumor cells were microdissected. Peptides from tumors, including eight cases of stage I and eight cases of stage II, were extracted and labeled with eight-channel iTRAQ reagents. The labeled peptides were identified and quantified by liquid chromatography–mass spectrometry (LC-MS) / MS using an LTQ Orbitrap Velos mass spectrometer. The candidate proteins were further evaluated by immunohistochemistry in lung cancer TMAs.
Results: A total of 1288 peptides (370 proteins) were identified and quantified. Among them, 173 proteins showed greater than a 1.5-fold difference between stage I and stage II tumors. Fifteen proteins showed significant changes in expression during tumor progression. The particularly interesting ones include fibrillin-2, annexin A5, myeloperoxidase, eukaryotic translation initiation factor 4A1 (eIF-4A1), and mucin-5B. Potential cellular functions of these proteins are summarized in Table 1.
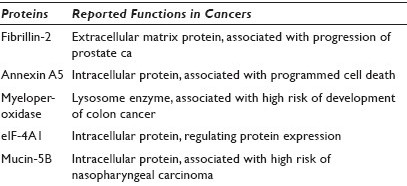
Conclusions: By using quantitative iTRAQ proteomic analysis, we are able to identify tumor proteins associated with early stage of lung adenocarcinoma. The significance of the differential expression of these staging-associated proteins is not well-studied in lung cancer, and they may be used as new potential protein biomarkers in both early detection of lung cancer and prediction of tumor progression. A further study is needed to validate these protein markers in lung cancers.
134: Assessment of nuclear nano-architecture characteristics for cervical cancer screening
Yang Liu, PhD1, Rajan Bista, PhD2, Shikhar Uttam, PhD2, Pin Wang, PhD2, Chengquan Zhao, MD3
1Departments of Medicine and Bioengineering, 2Medicine, 3Pathology, Magee-Womens Hospital, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania
Introduction: Cervical cancer is the second most common cancer in women worldwide. Papanicolaou (Pap) cytology is an established screening technique that has led to a significant reduction of incidence and mortality in cervical cancer. However, among the 55 million Pap cytology tests performed in the United States each year, only less than one percent of the patients have high-grade cervical intraepithelial lesions that require further extensive and invasive examination and treatment. Our recently developed novel optical technology — Spatial-domain Low-coherence Quantitative Phase Microscopy (SL-QPM) takes advantages of the ultra-sensitivity of the light interference effect and allows the detection of subtle structural alterations in the cell nuclei This technique is able to detect a minute change in nuclear structure, as small as 0.9 nm, on a scale ~1000 times smaller than what a conventional microscope can distinguish. This instrument can be directly used on the standard liquid-based Pap cytology slides without any modification. The goal of this pilot study is to evaluate the capability of SL-QPM-derived nuclear nano-architecture characteristics, to identify those patients who are likely to have high-grade squamous lesions on the follow-up colposcopy, in women with atypical Pap cytology diagnosis.
Materials and Methods: We performed SL-QPM on archived liquid-based Pap cytology specimens from 109 women (23 LSIL, 22 HSIL, 64 ASC-US) who underwent regular cervical cancer screening. Among those 64 ASC-US cases, 25 women had negative high-risk human papillomavirus (hrHPV) test (Hybrid Capture II) and 39 had positive hrHPV test (20 women had follow-up biopsies of CIN2 / 3). The slides were reviewed by a cytopathologist who marked the cells of interest for SL-QPM analysis. We analyzed the cell nuclei from approximately 15 to 20 cells per patient.
Results: A series of SL-QPM-derived nuclear nano-architecture were derived from both cytologically normal intermediate squamous cells and abnormal diagnostic cells, such as, average nuclear optical path length (a measure of nuclear density), intra-nuclear structural heterogeneity, and intra-nuclear entropy. For patients with HSIL and LSIL, these nuclear nano-architecture parameters were statistically different in normal, LSIL, and HSIL cells (P < 0.00001). For women with ASC-US cytology, they could distinguish cases with positive hrHPV from negative hrHPV with a very high level of statistical significance (P < 0.0001). More importantly, SL-QPM-derived nuclear nano-architecture parameters could distinguish those ASC-US cases with CIN2 / 3 on the follow-up biopsies with statistical significance (P < 0.05, 90% sensitivity and 74% specificity) from those ASC-US cases with positive hrHPV test without CIN2 / 3, in the follow-up biopsies.
Conclusions: The use of SL-QPM derived nuclear nano-architecture parameters represents a novel type of optical biomarker for cervical cancer screening. This optical biomarker could be incorporated into a diagnostic algorithm in which those patients with ASC-US Pap cytology would undergo SL-QPM analysis. Future studies are warranted to improve and evaluate this novel approach through the expansion of our study population and the identification of additional optical or other biomarkers to improve the prediction accuracy.
135: Uveal melanoma prognostication: Analysis of a method using fine needle aspiration and fluorescence in-situ hybridization
Abberly Lott Limbach, MD1, Arun Singh, MD3, Raymond Tubbs, DO2, Mary Turell, MD3, Dana Weber, CT(ASCP)1, Charles Biscotti, MD1
1Anatomic Pathology, Cleveland Clinic Foundation, Cleveland, Ohio; 2Molecular pathology, Cleveland Clinic Foundation, Cleveland, Ohio; 3Ophthalmology, Cleveland Clinic Foundation, Cleveland, Ohio
Introduction: Uveal melanoma patients can be segregated into two prognostic groups. Approximately half will develop metastases, while the other half will be cured with treatment of the uveal lesion. Unfortunately, the prognosis is cast by the time of diagnosis. Most experts agree that occult metastases exist at the time of primary diagnosis. The presence of monosomy 3 identifies the poor prognosis patients. Fluorescence in-situ hybridization (FISH) can identify monosomy 3. Unfortunately, FISH methods have varied greatly. Cytological samples are well suited for FISH analysis because they lack the artifacts associated with tissue sections. We have prospectively analyzed a series of uveal melanoma patients, in part, to determine the effectiveness of a prognostication technique that includes fine needle aspiration (FNA) and FISH analysis.
Materials and Methods: All patients had a clinical diagnosis of uveal melanoma based on the history and physical examination, including an ophthalmoscopic examination performed by one of the authors (ADS). The clinical assessment, most importantly the tumor size, determined whether the patients were treated with plaque radiotherapy or enucleation. FNA using a 25 G needle was performed at the time of plaque placement or enucleation by two of the authors (ADS and MET). In vivo samples were obtained by the transvitreal or transscleral approach, depending on the tumor location. Transvitreal samples were limited to a single pass. The aspirate samples were entirely rinsed into 2 – 3 ml of normal saline, visually examined, then submitted to cytology in CytoLyt (Hologic Corp, Marlborough, MA). ThinPrep® processing yielded four Papanicolaou stained slides per case. One of the authors (CVB) examined all the slides and depending on the cellularity, submitted up to three slides per case for FISH analysis. We counted CEP3 signals and used a 20% threshold for monosomy 3. Slides adequate for monosomy 3 evaluation had at least 200 cells counted.
Results: The 84 patients included 42 (50%) women and 42 men ranging in age from 26 to 91 years (mean 61 years). The tumors involved the choroid (59 cases, 70%), ciliochoroid (18 cases, 21%), iridociliary (4 cases, 4.8%), and iris (3 cases, 3.6%). Tumors ranged from 3.0 × 3.3 mm to 16 × 16 mm in basal dimensions and 1.2 mm to 15 mm in maximal height. FNA confirmed melanoma in 74 (88%) cases. Ten (12%) samples were insufficient for diagnosis. All diagnostic samples had melanoma cells well preserved for FISH analysis. Fifty-six (67%) patients had in vivo, transvitreal (55%) or transscleral (45%), aspirates. Thirty-seven (66%) of these had cellularity adequate for a 200 cell count FISH analysis. Twenty-eight (33%) patients had enucleation specimens aspirated and 26 (93%) of these had cellularity adequate for a 200 cell count FISH analysis. Overall, 43% of the tumors had monosomy 3.
Conclusions: Fine needle aspiration combined with FISH effectively identifies monosomy 3 in uveal melanomas. Overall, 88% of the cases in our series had diagnostic aspirate samples including cellularity adequate for a 200 cell count FISH analysis in 93% of the enucleation cases and 66% of the in vivo aspirate samples. The lower adequacy rate in the in vivo group is limiting, but not unexpected, as by definition, these tumors are smaller and thinner than those treated by enucleation.
136: Nanoscale nuclear architecture for urine cytology using spatial-domain low-coherence quantitative phase microscopy
Liron Pantanowitz, MD1, Rajan Bista, PhD2, Walid Khalbuss, MD, PhD1, Rajiv Dhir, MD1, Randall Brand, MD2, Sara Monaco, MD1, Yang Liu, PhD3
1Pathology, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania; 2Medicine, University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania; 3Bioengineering, University of Pittsburgh, Pittsburgh, Pennsylvania
Introduction: Definitive diagnosis of urothelial carcinoma in urine specimens based on cytomorphology alone may be challenging. Ancillary techniques such as fluorescence in-situ hybridization (FISH) have thus been employed to improve the sensitivity of cytology. We propose the use of a novel optical technique — spatial-domain low-coherence quantitative phase microscopy (SL-QPM) for analyzing nuclear architectural characteristics on the original unmodified cytology specimens with the goal of improving the sensitivity of urine cytology. This technique is able to detect a minute change in nuclear structure as small as 0.9 nm, a scale of about 1000 times smaller than what a conventional microscope detects. The aim of this study is to explore the ability of SL-QPM to improve the identification of malignant cells in urine cytology samples.
Materials and Methods: Urine samples were processed with ThinPrep® and submitted for FISH (Urovision). Urothelial cells from four benign (FISH-negative) and three positive (FISH-positive) cases for urothelial carcinoma were analyzed with SL-QPM, using Pap-stained ThinPrep® slides. The urothelial cells were classified into one of the three following scenarios: Benign cells in benign cases, morphologically benign-appearing cells in positive cases (i.e., uninvolved), and malignant cells in positive cases. SL-QPM-derived nanoscale nuclear architecture characteristics described by several optical parameters (i.e., optical path length, refractive index fluctuation ratio, intra-nuclear entropy, and uniformity) were analyzed from individual cell nuclei (4 – 25 cells / slide).
Results: All the SL-QPM-derived optical parameters were significantly different between morphologically benign and malignant urothelial cells (P = 5E-11). More importantly, the statistically significant differences for these optical parameters were also demonstrated among morphologically benign urothelial cells in both FISH-negative and FISH-positive cases (P = 0.03), as shown in Figure 1.
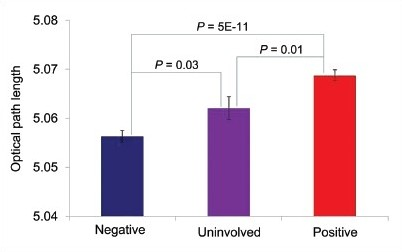
- Optical path length parameter shown for morphologically benign and malignant urothelial cells
Conclusions: We show that quantitative analysis of urothelial cell nuclei can be performed using original cytology slides without additional processing. Imaging of nanoscale nuclear architectural characteristics can readily identify malignant urothelial cells in Pap-stained urine specimens. The presence of nanoscale abnormalities in benign appearing cells from known cancer cases indicates a nuclear field defect that may not be evident by cytomorphology alone. This practical optical microscopy technique holds great promise as an ancillary technique to help improve the sensitivity of urine cytology of urothelial cancer and warrants further study.
137: Molecular diagnostics of metastatic melanoma utilizing cytological direct smears
Michael Roh, MD, PhD, Kim Hookim, MD, Joseph Willman, MD, Jeremiah Placido, MD, Helmut Weigelin, MLS(ASCP), Kristina Fields, BS, Judy Pang, MD, Bryan Betz, MD, Stewart Knoepp, MD
Pathology, University of Michigan Medical School, Ann Arbor, Michigan
Introduction: Activating mutations in the BRAF oncogene are present in at least 40% of the melanomas, with V600E and V600K representing the most commonly observed mutations. Clinical trials currently investigating the efficacy of targeted chemotherapy against mutant BRAF exemplify the heightened emphasis on personalized medical care. Hence, the use of fine needle aspiration (FNA) specimens of melanoma to assess BRAF mutations is increasing. Cell blocks are traditionally used for these studies. However, cell blocks exhibit variable cellularity, and in a significant proportion of cases, are of insufficient tumor cellularity. Direct smears represent an alternative platform for ancillary studies. Previously, we successfully demonstrated the application of immunocytochemistry as well as the epidermal growth factor receptor (EGFR) and KRAS mutational analyses to the cytological direct smears prepared from FNAs of non-small cell lung carcinomas. In this study, we investigate the application of immunocytochemistry and BRAF mutational analysis to direct smears prepared from FNAs of metastatic melanoma.
Materials and Methods: First, immunocytochemistry for S-100, HMB45, and Mart-1 was prospectively performed on air-dried, unstained, direct smears following brief formalin fixation (30 – 60 minutes), and also antigen retrieval in 16 consecutive FNAs of metastatic melanoma. Next, in 15 additional cases obtained from the archive, BRAF sequencing was performed using genomic DNA isolated from microdissected tumor cell–enriched areas on de-coverslipped, Diff Quik-stained smears. In parallel, BRAF sequencing was performed using DNA extracted from the corresponding cell blocks.
Results: Of the 16 melanoma FNAs in which immunocytochemistry was performed on unstained direct smears, S-100 positivity in the tumor cells was observed in all cases. HMB45 and Mart-1 positivity was seen in 80 and 88% of the cases, respectively. All three markers were positive in 75% of the cases [Figure 1]. Next, for the 15 archived melanoma FNAs, the cellular material microdissected from Diff-Quik® stained smears [Figure 2] yielded high quality genomic DNA in all cases. Mutations in BRAF were observed at the expected frequency; eight (53%) of the fifteen cases tested positive. Five and three melanomas harbored the V600E and V600K mutations, respectively [Figure 3]. Concordant sequencing results for cellular material obtained from direct smears and cell blocks were observed in 14 (93%) of the 15 cases; in one case, the mutation was not detected in the cell block material, whereas, the V600E mutation was detected in material obtained from the direct smear. There were no cases in which a BRAF mutation was detected in the cell block material, but not in the cellular material from the smears.
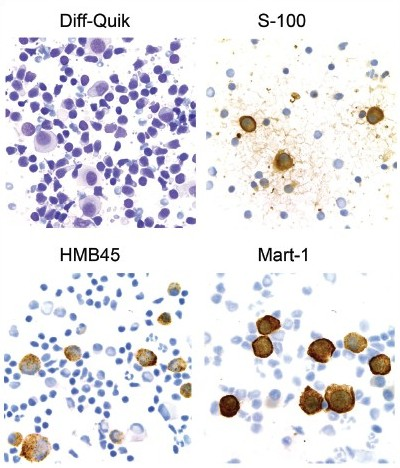
- Application of immunocytochemistry to direct smears of metastatic melanoma
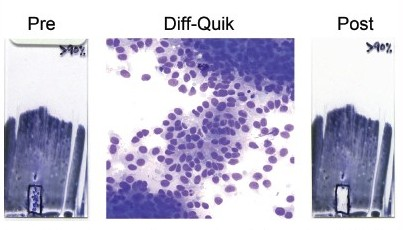
- Microdissection of tumor cell–enriched areas from Diff-Quik® stained direct smears of melanoma
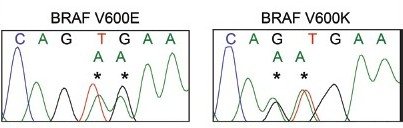
- BRAF mutations detected in cellular material obtained from cytological direct smears
Conclusions: This study demonstrates that direct smears represent a robust and valuable source of cellular material for ancillary studies utilized in the cytological diagnosis of melanoma. Unstained, air-dried smears can be effectively utilized for confirmatory immunocytochemical studies. Furthermore, Diff-Quik® stained smears allow for direct visualization and microdissection of tumor-enriched areas for DNA isolation and subsequent BRAF mutation analysis. The application of these ancillary studies to cytological smears can be effectively applied, especially in scenarios when inadequate cell block cellularity is anticipated or encountered.
138: Emerin: A useful immunohistochemical marker for highlighting nuclear membrane irregularities in papillary thyroid carcinoma
Mary Kinsella, Cynthia Cohen, Momin Siddiqui
Pathology and Laboratory Medicine, Emory University Hospital, Atlanta, Georgia
Introduction: The basic cytological diagnosis of papillary thyroid carcinoma (PTC) and its distinction from follicular neoplasia (FN) relies on a constellation of sometimes subtle morphological changes in the nucleus, including shape, chromatin distribution, and the presence of intra-nuclear inclusions and grooves. Immunohistochemical staining for Emerin, an integral inner nuclear membrane protein, has recently been shown to highlight fine nuclear membrane irregularities and structural changes in PTC that are not apparent with conventional H and E staining. The purpose of this study is to examine the diagnostic utility of emerin IHC in the cytological preparations of PTC and FN.
Materials and Methods: Alcohol- or formalin-fixed, paraffin-embedded cell blocks from FNAs of PTC or FN were stained with anti-Emerin polyclonal antibody and were subsequently evaluated for nuclear grooves, circumferential nuclear membrane irregularities (garlands), inclusions, deep stellate membrane invaginations, crescents, and nuclear shape variation. Additional clinical and pathological data, including patient gender and age, tumor size, and final histological diagnosis, were also obtained.
Results: A total of 34 FNA cases were evaluated, 24 were cytologically diagnosed as PTC and 10 as FN (three follicular variants of PTC on surgical follow-up). The final total number of PTC cases examined was 27, with seven FN. Emerin IHC of PTC revealed a predominantly oval nuclear shape in a majority of the cases (88%), with FN demonstrating round nuclei and FV of PTC showing a roughly equal distribution of round and oval shapes. In addition to the oval nuclear shape (P = 0.004), the presence of Emerin-positive nuclear grooves (P < 0.001), circumferential Emerin nuclear garlands (P = 0.006), and chromatin clearing on H and E staining (P = 0.02), in combination, were significant predictors of PTC by regression analysis. The average age of the patients was 49 years, and the average tumor size was 2.5 cm.
Conclusions: Emerin IHC of thyroid FNA specimens serves as a useful adjunct to conventional H and E staining in the diagnosis of PTC and its distinction from FN, by delineating the diagnostic nuclear membrane irregularities (garlands), nuclear grooves, and a characteristic oval nuclear shape. For diagnostically challenging cases with limited cellularity, emerin staining can help in providing a definitive diagnosis of PTC.
139: Methylation of O6-methylguanine-DNA methyltransferase as a chemosensitive biomarker in glioblastoma in fine needle aspiration biopsy samples
Bin Yang, MD, PhD, Rosemarie Read, PhD, Raymond Tubbs, DO
Pathology, Cleveland Clinic, Cleveland, Ohio
Introduction: O6-Methylguanine-DNA methyltransferase (MGMT) is a DNA repair enzyme that specifically removes promutagenic alkyl groups from the O6 position of guanine in DNA. Repair of O6-alkylguanine adducts by tumor cells has been implicated in drug resistance, as it reduces the cytotoxicity of alkylating chemotherapeutic agents. Recent studies indicate that patients with MGMT methylation have a better response to temozolomide and their median survival nearly doubles compared to those with lack of MGMT methylation. In order to provide this molecular biomarker clinically in triage patients for chemotherapy, we have developed and validated the MGMT methylation assay with pyrosequencing on the Fine Needle Aspiration Biopsy (FNAB) samples of glioblastoma.
Materials and Methods: Promoter methylation of MGMT was quantitatively analyzed by Pyro Q96. MGMT methylation was tested and validated with 33 cases of glioblastoma and 10 cases of non-neoplastic brain tissue. The analytical sensitivity and minimal requirement of the DNA amount was evaluated with two cell lines, a cancer cell line harboring of MGMT methylation and a normal cell line with lack of MGMT methylation. The analytical sensitivity between pyrosequencing and methylation-specific polymerase chain reaction (PCR) is also compared.
Results: Methylation of MGMT was identified in 33% (11 / 33) of the cases of glioblastoma and none in the epilepsy brain tissue. The average methylation level of CpG islands in the MGMT promoter was > 30% in glioblastoma and < 3% in epilepsy brain. By a series dilution of a methylated cancer cell line with an unmethylated normal cell line, the analytical sensitivity for pyrosequencing detection of MGMT methylation was at 5%. The minimal amount of genomic DNA required to successfully detect MGMT methylation by pyrosequencing was at 100 ng (approximately 3,000 cells). In comparison with MSP, pyrosequencing was comparably sensitive with less false-positive cases and also provided a quantitative methylation value for each CpG island.
Conclusions: We have validated the MGMT methylation assay clinically in providing a powerful biomarker for triaging patients with glioblastoma for chemotherapy. This assay can be performed both on FNAB tissue blocks and on cytological smears. We demonstrate that pyrosequencing detection of MGMT methylation has an analytical sensitivity suitable for clinical utility.








